8QTN
 
 | | Cryo-EM structure of the apo yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM Structure of the Yeast Ceramide Synthase Complex
To Be Published
|
|
8QTR
 
 | | Cryo-EM structure of the FB-bound yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Yeast Ceramide Synthase Complex
To Be Published
|
|
8QOF
 
 | | Cryo-EM structure of the yeast SPT-Orm2-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
8QOG
 
 | | Cryo-EM structure of the yeast SPT-Orm2-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
8C80
 
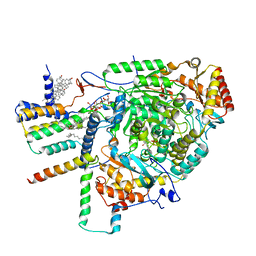 | | Cryo-EM structure of the yeast SPT-Orm1-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C81
 
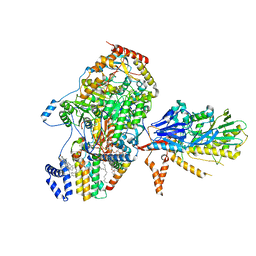 | | Cryo-EM structure of the yeast SPT-Orm1-Sac1 complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C82
 
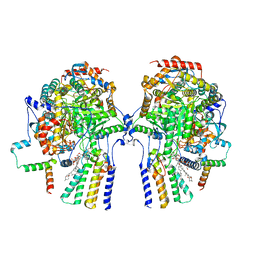 | | Cryo-EM structure of the yeast SPT-Orm1-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C7G
 
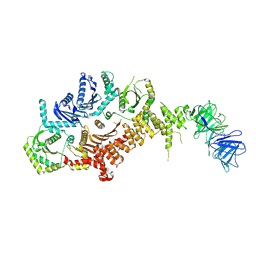 | | Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli | | Descriptor: | Caffeine, calcium, zinc sensitivity 1, ... | | Authors: | Schaefer, J, Herrmann, E, Kuemmel, D, Moeller, A. | | Deposit date: | 2023-01-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6S9T
 
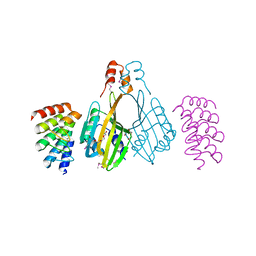 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 3 | | Descriptor: | Darpin 3, LIM domain-binding protein 1, TETRAETHYLENE GLYCOL | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S9S
 
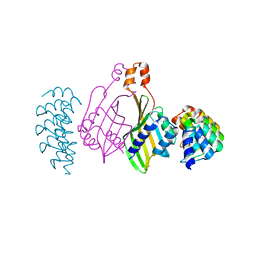 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | Descriptor: | Darpin 10, LIM domain-binding protein 1 | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7B4T
 
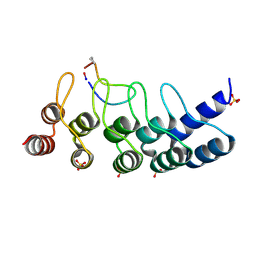 | | Broadly neutralizing DARPin bnD.1 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.1, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), SULFATE ION | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4U
 
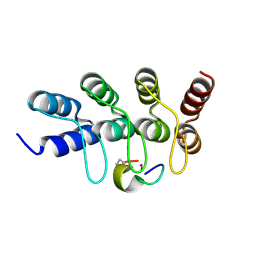 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CALCIUM ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4V
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CHLORIDE ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4W
 
 | | Broadly neutralizing DARPin bnD.3 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Broadly neutralizing DARPin bnD.3, ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
6FP8
 
 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPA
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP7
 
 | | mTFP1/DARPin 1238_E11 complex in space group P6522 | | Descriptor: | DARPin1238_E11, GFP-like fluorescent chromoprotein cFP484, GLYCEROL | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPB
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
3H1Z
 
 | | Molecular basis for the association of PIPKIgamma -p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit beta-1, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Krainer, G, Keller, S, Saenger, W, Krauss, M, Haucke, V. | | Deposit date: | 2009-04-14 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
3H85
 
 | | Molecular basis for the association of PIPKI gamma-p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit mu-1, NICKEL (II) ION, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Haucke, V. | | Deposit date: | 2009-04-28 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
3HS9
 
 | | Intersectin 1-peptide-AP2 beta ear complex | | Descriptor: | AP-2 complex subunit beta-1, peptide from Intersectin-1, residues 841-851 | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HS8
 
 | | Intersectin 1-peptide-AP2 alpha ear complex | | Descriptor: | Adaptor protein complex AP-2, alpha 2 subunit, peptide from Intersectin-1, ... | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
