3GHY
 
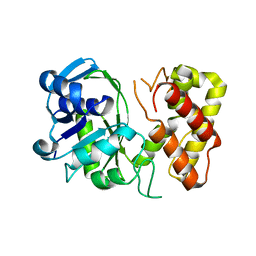 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3IJ6
 
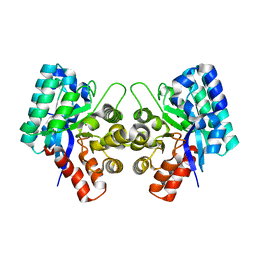 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | Descriptor: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
3H12
 
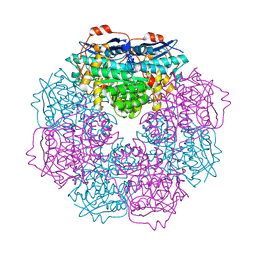 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
3GT7
 
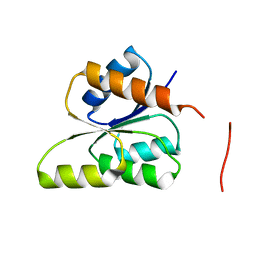 | | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF SIGNAL TRANSDUCTION HISTIDINE KINASE FROM Syntrophus aciditrophicus | | Descriptor: | Sensor protein | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Signal Transduction Kinase from Syntrophus Aciditrophicus
To be Published
|
|
3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
3HQC
 
 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3HV2
 
 | | Crystal structure of signal receiver domain OF HD domain-containing protein FROM Pseudomonas fluorescens Pf-5 | | Descriptor: | Response regulator/HD domain protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of signal receiver domain oF HD domain-containing protein 3 FROM Pseudomonas fluorescens
To be Published
|
|
3IJD
 
 | | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
3IRV
 
 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
3ISA
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase/isomerase FROM Bordetella parapertussis | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative enoyl-CoA hydratase/isomerase | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF enoyl-CoA hydratase FROM Bordetella parapertussis
To be Published
|
|
3IWA
 
 | | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris | | Descriptor: | CALCIUM ION, FAD-dependent pyridine nucleotide-disulphide oxidoreductase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris
To be Published
|
|
3IAN
 
 | | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, SODIUM ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis
To be Published
|
|
3IRS
 
 | | CRYSTAL STRUCTURE OF UNCHARACTERIZED TIM-BARREL PROTEIN BB4693 FROM Bordetella bronchiseptica | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Do, J, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF UNCHARACTERIZED HYDROLASE FROM Bordetella bronchiseptica
To be Published
|
|
3IBM
 
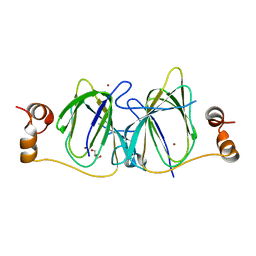 | | CRYSTAL STRUCTURE OF cupin 2 domain-containing protein Hhal_0468 FROM Halorhodospira halophila | | Descriptor: | Cupin 2, conserved barrel domain protein, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF cupin 2 domain-containing PROTEIN Hhal_0468 FROM Halorhodospira halophila
To be Published
|
|
3ICJ
 
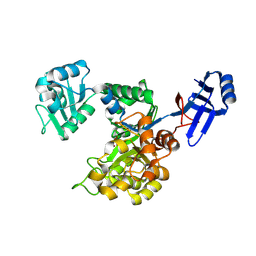 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | Descriptor: | ZINC ION, uncharacterized metal-dependent hydrolase | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
3I76
 
 | | The crystal structure of the orthorhombic form of the putative HAD-hydrolase YfnB from Bacillus subtilis bound to magnesium reveals interdomain movement | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the orthorhombic form of the putative HAD-hydrolase YfnB from Bacillus subtilis bound to magnesium reveals interdomain movement
To be Published
|
|
3GY1
 
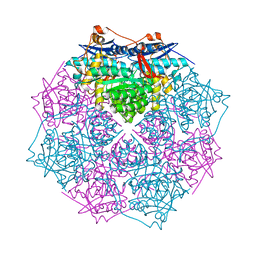 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3GO2
 
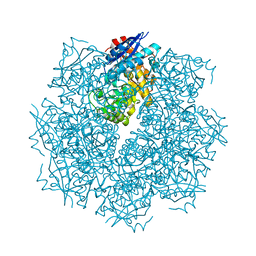 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium | | Descriptor: | MAGNESIUM ION, Putative L-alanine-DL-glutamate epimerase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium.
To be Published
|
|
3GZ4
 
 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3I5T
 
 | | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides KD131 | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides
To be Published
|
|
3I47
 
 | | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Enoyl CoA hydratase/isomerase (Crotonase) | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila
subsp. pneumophila str. Philadelphia 1
To be Published
|
|
3I4K
 
 | | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum
To be Published
|
|
3IGH
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus horikoshii ot3 | | Descriptor: | SULFATE ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Horikoshii
To be Published
|
|
3IMH
 
 | | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus NCFM | | Descriptor: | CHLORIDE ION, GLYCEROL, Galactose-1-epimerase, ... | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus
To be Published
|
|
