2N37
 
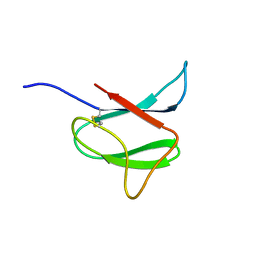 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
3TTI
 
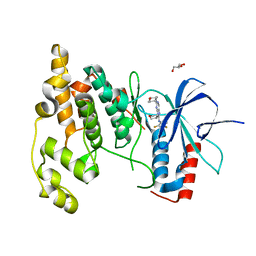 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TTJ
 
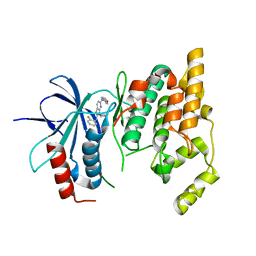 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1WZZ
 
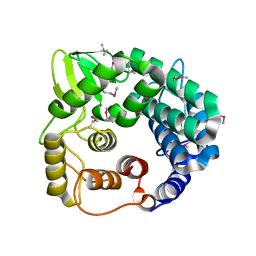 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
3WVQ
 
 | | Structure of ATP grasp protein | | Descriptor: | GLYCEROL, PGM1, SULFATE ION | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
3WVR
 
 | | Structure of ATP grasp protein with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PGM1, ... | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
