4G1Y
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | (4E,6Z)-7-(3-{[3,4-bis(hydroxymethyl)benzyl]oxy}phenyl)-3-ethylnona-4,6-dien-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G1Z
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G2H
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4K2A
 
 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | 分子名称: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | 登録日 | 2013-04-08 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G2I
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Vitamin D3 receptor | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
2V9Z
 
 | | Structure of the Rhodococcus haloalkane dehalogenase mutant with enhanced enantioselectivity | | 分子名称: | HALOALKANE DEHALOGENASE | | 著者 | Koudelakova, T, Prokop, Z, Sato, Y, Lapkouski, M, Chovancova, E, Monincova, M, Jesenska, A, Emmer, J, Senda, T, Nagata, Y, Kuta Smatanova, I, Damborsky, J. | | 登録日 | 2007-08-28 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Rational Engineering of Rhodococcus Haloalkane Dehalogenase with Enhanced Enantioselectivity
To be Published
|
|
5GTG
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing 1,2-propanediol | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lachrymatory-factor synthase, S-1,2-PROPANEDIOL, ... | | 著者 | Arakawa, T, Sato, Y, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | 登録日 | 2016-08-20 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
3D24
 
 | | Crystal structure of ligand-binding domain of estrogen-related receptor alpha (ERRalpha) in complex with the peroxisome proliferators-activated receptor coactivator-1alpha box3 peptide (PGC-1alpha) | | 分子名称: | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR1 | | 著者 | Moras, D, Greschik, H, Flaig, R, Sato, Y, Rochel, N, Structural Proteomics in Europe (SPINE) | | 登録日 | 2008-05-07 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Communication between the ERR{alpha} Homodimer Interface and the PGC-1{alpha} Binding Surface via the Helix 8-9 Loop.
J.Biol.Chem., 283, 2008
|
|
5GVC
 
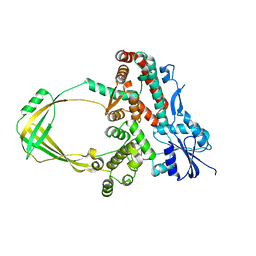 | | Human Topoisomerase IIIb topo domain | | 分子名称: | DNA topoisomerase 3-beta-1, MAGNESIUM ION | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | 登録日 | 2016-09-05 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.436 Å) | | 主引用文献 | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVD
 
 | | Human TDRD3 DUF1767-OB domains | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | 登録日 | 2016-09-05 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.623 Å) | | 主引用文献 | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVE
 
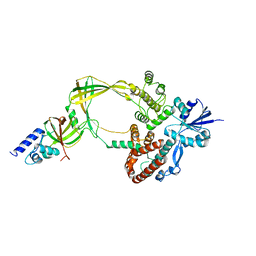 | | Human TOP3B-TDRD3 complex | | 分子名称: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | 登録日 | 2016-09-05 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.606 Å) | | 主引用文献 | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
7VNN
 
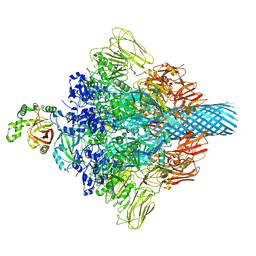 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
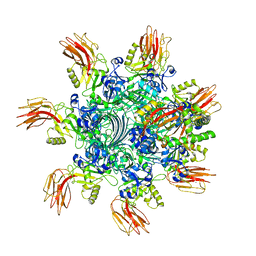 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
5B86
 
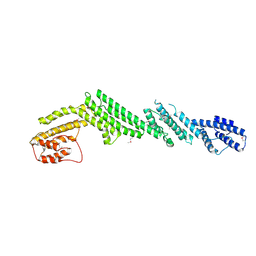 | | Crystal structure of M-Sec | | 分子名称: | Tumor necrosis factor alpha-induced protein 2 | | 著者 | Yamashita, M, Sato, Y, Yamagata, A, Fukai, S. | | 登録日 | 2016-06-12 | | 公開日 | 2016-10-12 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (3.017 Å) | | 主引用文献 | Distinct Roles for the N- and C-terminal Regions of M-Sec in Plasma Membrane Deformation during Tunneling Nanotube Formation.
Sci Rep, 6, 2016
|
|
5XNY
 
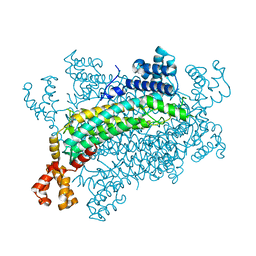 | | Crystal structure of CreD | | 分子名称: | CreD | | 著者 | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | 登録日 | 2017-05-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XNZ
 
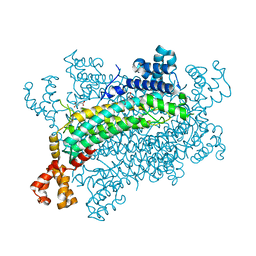 | | Crystal structure of CreD complex with fumarate | | 分子名称: | CreD, FUMARIC ACID | | 著者 | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | 登録日 | 2017-05-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
6IXF
 
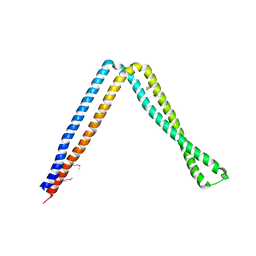 | |
6IXE
 
 | | Crystal structure of SeMet apo SH3BP5 (I41) | | 分子名称: | SH3 domain-binding protein 5, SUCCINIC ACID | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2018-12-10 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6IXV
 
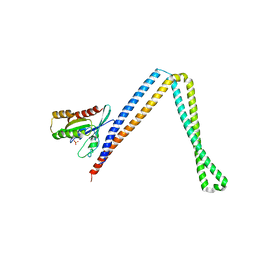 | | Crystal structure of SH3BP5-Rab11a | | 分子名称: | PHOSPHATE ION, Ras-related protein Rab-11A, SH3 domain-binding protein 5 | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2018-12-12 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6IXG
 
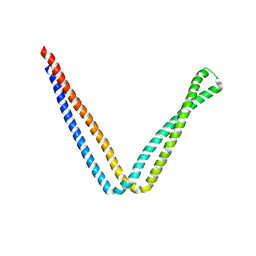 | |
4Y61
 
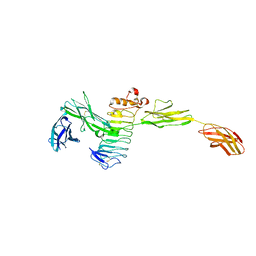 | | Crystal structure of the complex between Slitrk2 LRR1 and PTP delta Ig1-Fn1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 2 | | 著者 | Yamgata, A, Sato, Y, Goto-Ito, S, Uemura, T, Maeda, A, Shiroshima, T, Yoshida, T, Fukai, S. | | 登録日 | 2015-02-12 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.358 Å) | | 主引用文献 | Structure of Slitrk2-PTP delta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion.
Sci Rep, 5, 2015
|
|
5YDK
 
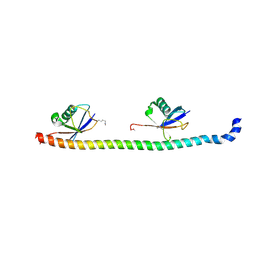 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, tetrameric form | | 分子名称: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, Ubiquitin-40S ribosomal protein S27a | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168
Nat Commun, 9, 2018
|
|
5YOY
 
 | | Crystal structure of the human tumor necrosis factor in complex with golimumab Fv | | 分子名称: | Golimumab heavy chain variable region, Golimumab light chain variable region, Tumor necrosis factor | | 著者 | Ono, M, Horita, S, Sato, Y, Nomura, Y, Iwata, S, Nomura, N. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.727 Å) | | 主引用文献 | Structural basis for tumor necrosis factor blockade with the therapeutic antibody golimumab
Protein Sci., 27, 2018
|
|
5XIT
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | 分子名称: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | 分子名称: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
