5A8E
 
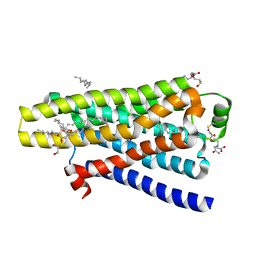 | | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-[(2S)-3-(tert-butylamino)-2-hydroxypropoxy]-7-methyl-1H-indole-2-carbonitrile, ... | | Authors: | Sato, T, Baker, J.G, Warne, T, Brown, G.A, Congreve, M, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor.
Mol.Pharmacol., 88, 2015
|
|
1ETL
 
 | |
1ETM
 
 | |
3VLH
 
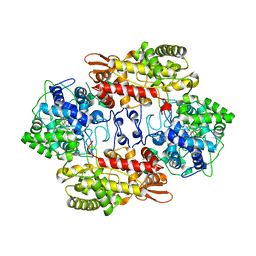 | |
3VLI
 
 | |
3VLJ
 
 | | Crystal Structure Analysis of the Cyanide Arg409Leu Variant Complexes with o-Dianisidine in KatG from HALOARCULA MARISMORTUI | | Descriptor: | 3,3'-dimethoxybiphenyl-4,4'-diamine, CYANIDE ION, Catalase-peroxidase 2, ... | | Authors: | Sato, T, Higuchi, W, Yoshimatsu, K, Fujiwara, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures and Peroxidatic Function of Cyanide Arg409Leu Variant and its Complexes with o-Dianisidine in KatG from HALOARCULA MARISMORTUI
To be Published
|
|
3VLK
 
 | |
1ETN
 
 | |
3VLM
 
 | | Crystal Structure Analysis of the Met244Ala Variant of KatG from Haloarcula marismortui | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sato, T, Ten-i, T, Higuchi, W, Yoshimatsu, K, Fujiwara, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure and Kinetic Studies on Met244Ala Variant of KatG from HALOARCULA MARISMORTUI
To be Published
|
|
3UW8
 
 | |
2A70
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Z
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 1 | | Descriptor: | Emp47p (form2) | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6W
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6X
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), Y131F mutant | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
3VLL
 
 | |
2CMM
 
 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
2D3Q
 
 | |
5JQP
 
 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | Authors: | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
5H18
 
 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
5DKZ
 
 | | Crystal structure of glucosidase II alpha subunit (alpha3-Glc2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DKX
 
 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DL0
 
 | | Crystal structure of glucosidase II alpha subunit (Glc1Man2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
