3AEI
 
 | | Crystal structure of the prefoldin beta2 subunit from Thermococcus strain KS-1 | | Descriptor: | CHLORIDE ION, Prefoldin beta subunit 2, SULFATE ION | | Authors: | Ohtaki, A, Sugano, Y, Sato, T, Noguchi, K, Miyatake, H, Yohda, M. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic Characterization of the Interaction between Prefoldin and Group II Chaperonin
J.Mol.Biol., 399, 2010
|
|
3VR0
 
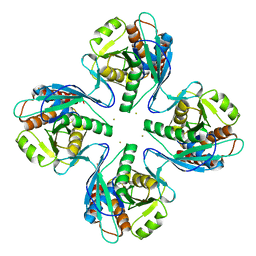 | | Crystal structure of Pyrococcus furiosus PbaB, an archaeal proteasome activator | | Descriptor: | GOLD ION, Putative uncharacterized protein | | Authors: | Kumoi, K, Satoh, T, Hiromoto, T, Mizushima, T, Kamiya, Y, Noda, M, Uchiyama, S, Murata, K, Yagi, H, Kato, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An archaeal homolog of proteasome assembly factor functions as a proteasome activator
Plos One, 8, 2013
|
|
8IC9
 
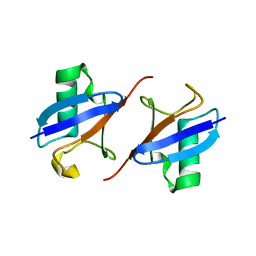 | | Lys48-linked K48C-diubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin | | Authors: | Hiranyakorn, M, Yagi-Utsumi, M, Yanaka, S, Ohtsuka, N, Momiyama, N, Satoh, T, Kato, K. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 24, 2023
|
|
5WTQ
 
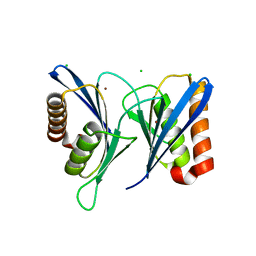 | | Crystal structure of human proteasome-assembling chaperone PAC4 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Proteasome assembly chaperone 4 | | Authors: | Kurimoto, E, Satoh, T, Ito, Y, Ishihara, E, Tanaka, K, Kato, K. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation
Protein Sci., 26, 2017
|
|
3VQR
 
 | | Structure of a dye-linked L-proline dehydrogenase mutant from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
7DBT
 
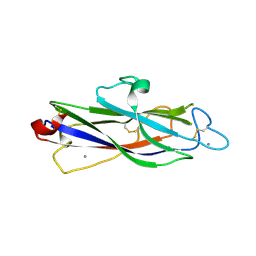 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Mn2+-bound form) | | Descriptor: | AMNP/g12777, MANGANESE (II) ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
7DBU
 
 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Zn2+-bound form) | | Descriptor: | AMNP/g12777, ZINC ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
3VYJ
 
 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 (apo form) | | Descriptor: | C-type lectin domain family 4, member a4, SULFATE ION | | Authors: | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
5BW7
 
 | | Crystal structure of nonfucosylated Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isoda, Y, Yagi, H, Satoh, T, Shibata-Koyama, M, Masuda, K, Satoh, M, Kato, K, Iida, S. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with Fc gamma RIIIa and Other Fc gamma Receptors
Plos One, 10, 2015
|
|
7CAP
 
 | | Cyclic Lys48-linked triubiquitin | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Hiranyakorn, M, Yanaka, S, Satoh, T, Wilasri, T, Jityuti, B, Yagi-Utsumi, T, Kato, K. | | Deposit date: | 2020-06-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 21, 2020
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
2RUF
 
 | |
2RUE
 
 | |
3WV0
 
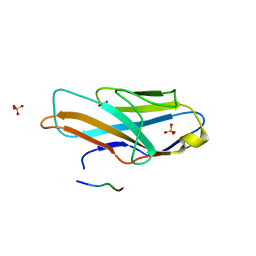 | | O-glycan attached to herpes simplex virus type 1 glycoprotein gB is recognized by the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | Envelope glycoprotein B, N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-galactopyranose, Paired immunoglobulin-like type 2 receptor alpha, ... | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WUZ
 
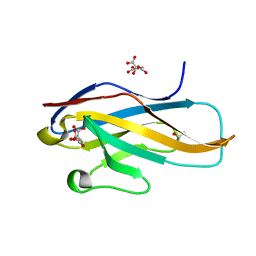 | | Crystal structure of the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WZS
 
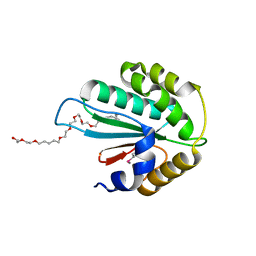 | | Crystal structure of Trx3 domain of UGGT (detergent-bound form) | | Descriptor: | 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Zhu, T, Satoh, T, Kato, K. | | Deposit date: | 2014-10-03 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into substrate recognition by the endoplasmic reticulum folding-sensor enzyme: crystal structure of third thioredoxin-like domain of UDP-glucose:glycoprotein glucosyltransferase
Sci Rep, 4, 2014
|
|
3WZT
 
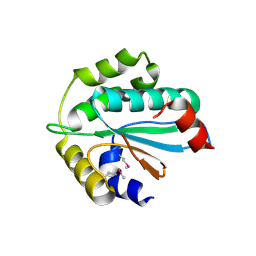 | | Crystal structure of Trx3 domain of UGGT (detergent-unbound form) | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Zhu, T, Satoh, T, Kato, K. | | Deposit date: | 2014-10-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into substrate recognition by the endoplasmic reticulum folding-sensor enzyme: crystal structure of third thioredoxin-like domain of UDP-glucose:glycoprotein glucosyltransferase
Sci Rep, 4, 2014
|
|
5CRW
 
 | |
5Y9N
 
 | | Crystal structure of Pyrococcus furiosus PbaA (monoclinic form), an archaeal homolog of proteasome-assembly chaperone | | Descriptor: | CHLORIDE ION, PbaA | | Authors: | Yagi-Utsumi, M, Sikdar, A, Kozai, T, Inoue, R, Sugiyama, M, Uchihashi, T, Satoh, T, Kato, K. | | Deposit date: | 2017-08-26 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conversion of functionally undefined homopentameric protein PbaA into a proteasome activator by mutational modification of its C-terminal segment conformation
Protein Eng. Des. Sel., 31, 2018
|
|
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
5XJE
 
 | | Crystal structure of fucosylated IgG1 Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5XJF
 
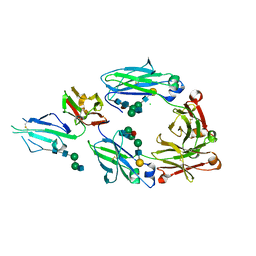 | | Crystal structure of fucosylated IgG Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
3VYK
 
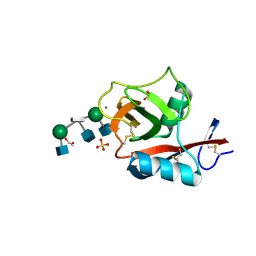 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 in complex with N-glycan | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, C-type lectin domain family 4, ... | | Authors: | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
3WT1
 
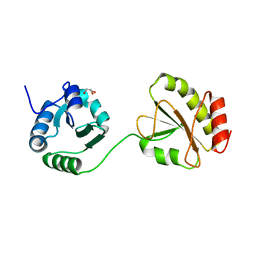 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (reduced form) | | Descriptor: | GLYCEROL, Protein disulfide-isomerase | | Authors: | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | Deposit date: | 2014-04-02 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
