7JZ0
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6AZI
 
 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6B8D
 
 | | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
To Be Published
|
|
6BAL
 
 | | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CHLORIDE ION, Malate dehydrogenase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate
To Be Published
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
6BXG
 
 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | Descriptor: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
6C4Q
 
 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
6BWT
 
 | | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, SULFATE ION, Thioredoxin reductase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis.
To Be Published
|
|
6C4V
 
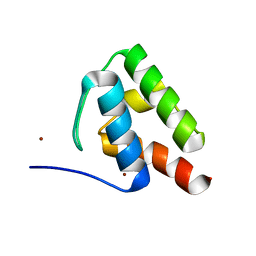 | | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | Polyketide synthase Pks13, ZINC ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To be Published
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | Descriptor: | Gamma-aminobutyraldehyde dehydrogenase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-11 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
6CN0
 
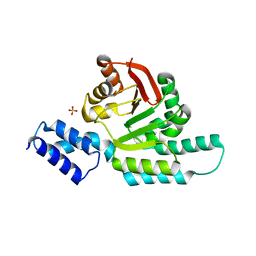 | | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Evdokimova, E, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis.
To Be Published
|
|
6CMZ
 
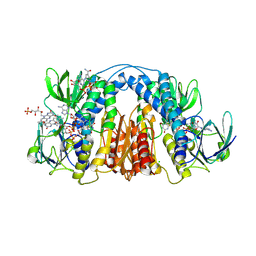 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
6D0P
 
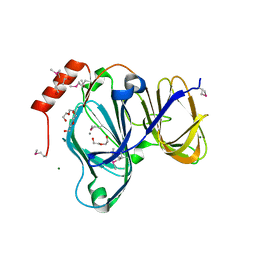 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
6U7L
 
 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6PI9
 
 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
6UOF
 
 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
6VBB
 
 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6VBF
 
 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
