5YPY
 
 | | Crystal structure of IlvN. Val-1c | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-04 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YUM
 
 | | Crystallographic structures of IlvN.Val/Ile complexes:Conformational selectivity for feedback inhibition of AHASs | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, COBALT (II) ION, ISOLEUCINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-22 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YPP
 
 | | Crystal structure of IlvN.Val-1a | | Descriptor: | ACETATE ION, Acetolactate synthase isozyme 1 small subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YPW
 
 | | Crystal structure of IlvN.Val-1b | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
6KNO
 
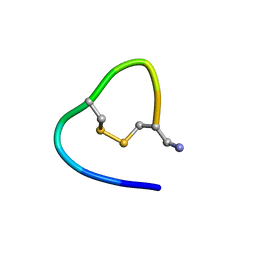 | |
6KMY
 
 | | Structure of single disulfide peptide Czon1107-P5A | | Descriptor: | Czon1107-P5A | | Authors: | Sarma, S.P, Madhan Kumar, M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and allosteric activity of a single-disulfide conopeptide fromConus zonatusat human alpha 3 beta 4 and alpha 7 nicotinic acetylcholine receptors.
J.Biol.Chem., 295, 2020
|
|
6KNP
 
 | |
6KN3
 
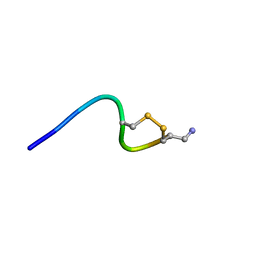 | |
6KN2
 
 | |
2MW7
 
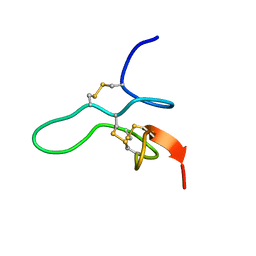 | |
2M61
 
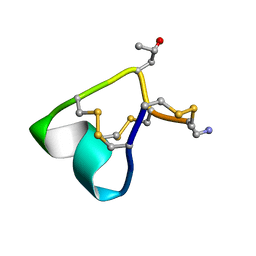 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
2M62
 
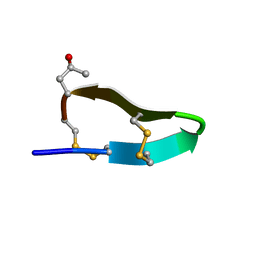 | |
1YZ2
 
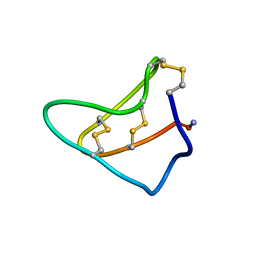 | | Solution structure of Am2766 | | Descriptor: | Delta-conotoxin Am 2766 | | Authors: | Sarma, S.P, Kumar, G.S, Sudarslal, S, Iengar, P, Sikdar, S.K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2005-02-26 | | Release date: | 2006-02-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of delta-Am2766: A Highly Hydrophobic delta-Conotoxin from Conus amadis That Inhibits Inactivation of Neuronal Voltage-Gated Sodium Channels
CHEM.BIODIVERS., 2, 2005
|
|
2KTC
 
 | | Solution Structure of a Novel hKv1.1 inhibiting scorpion toxin from Mesibuthus tamulus | | Descriptor: | Potassium channel toxin alpha-KTx 9.4 | | Authors: | Kumar, G.S, Upadhyay, S, Mathew, M.K, Sarma, S.P. | | Deposit date: | 2010-01-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BTK-2, a novel hK(v)1.1 inhibiting scorpion toxin, from the eastern Indian scorpion Mesobuthus tamulus.
Biochim.Biophys.Acta, 1814, 2011
|
|
8K3N
 
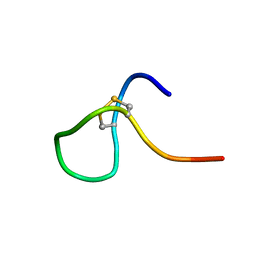 | |
8K3M
 
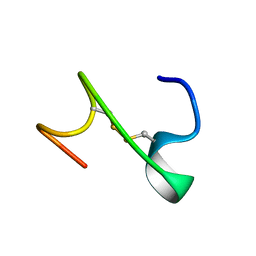 | |
7WJ0
 
 | |
7WNR
 
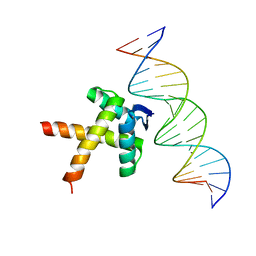 | | Data-driven HADDOCK model of mycobacterial nMazE6-operator DNA complex | | Descriptor: | Antitoxin MazE6, DNA (5'-D(P*CP*CP*GP*GP*TP*TP*AP*TP*AP*CP*TP*AP*TP*CP*TP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*AP*GP*AP*TP*AP*GP*TP*AP*TP*AP*AP*CP*CP*GP*G)-3') | | Authors: | Kumari, K, Sarma, S.P. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and mutational analysis of MazE6-operator DNA complex provide insights into autoregulation of toxin-antitoxin systems.
Commun Biol, 5, 2022
|
|
8YUM
 
 | |
8YSY
 
 | |
1B5A
 
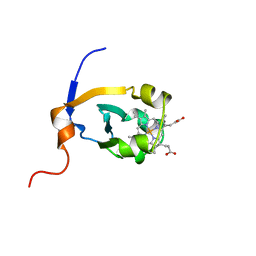 | | RAT FERROCYTOCHROME B5 A CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1B5B
 
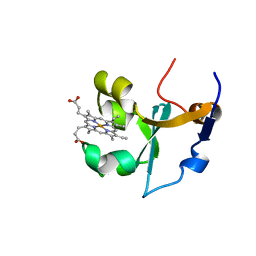 | | RAT FERROCYTOCHROME B5 B CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1KEB
 
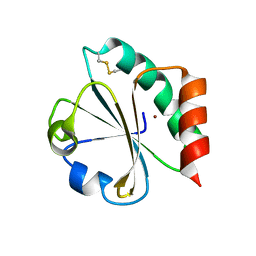 | | Crystal Structure of Double Mutant M37L,P40S E.coli Thioredoxin | | Descriptor: | COPPER (II) ION, Thioredoxin 1 | | Authors: | Rudresh, Jain, R, Dani, V, Mitra, A, Srivastava, S, Sarma, S.P, Varadarajan, R, Ramakumar, S. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Consequences of Replacement of an alpha-helical Pro Residue in E.coli Thioredoxin
PROTEIN ENG., 15, 2002
|
|
2FQ2
 
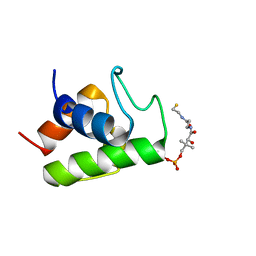 | | Solution structure of minor conformation of holo-acyl carrier protein from malaria parasite plasmodium falciparum | | Descriptor: | 4'-PHOSPHOPANTETHEINE, acyl carrier protein | | Authors: | Sharma, A.K, Sharma, S.K, Surolia, A, Surolia, N, Sarma, S.P. | | Deposit date: | 2006-01-17 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs
Biochemistry, 45, 2006
|
|
2FQ0
 
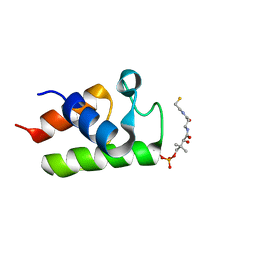 | | Solution structure of major conformation of holo-acyl carrier protein from malaria parasite plasmodium falciparum | | Descriptor: | 4'-PHOSPHOPANTETHEINE, acyl carrier protein | | Authors: | Sharma, A.K, Sharma, S.K, Surolia, A, Surolia, N, Sarma, S.P. | | Deposit date: | 2006-01-17 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs
Biochemistry, 45, 2006
|
|
