5JUV
 
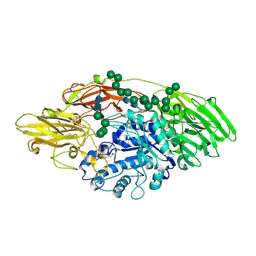 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
7ZY9
 
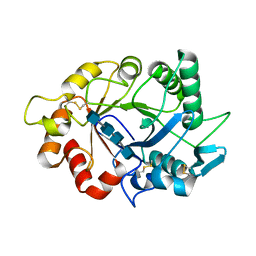 | |
7ZYA
 
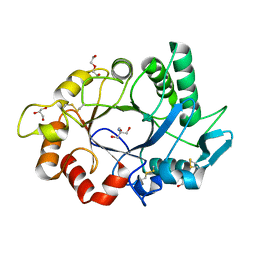 | | Structure of Chit33 from Trichoderma harzianum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Endochitinase 33, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure-Function Insights into the Fungal Endo -Chitinase Chit33 Depict its Mechanism on Chitinous Material.
Int J Mol Sci, 23, 2022
|
|
7PN6
 
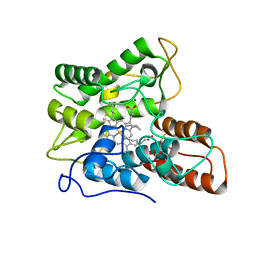 | |
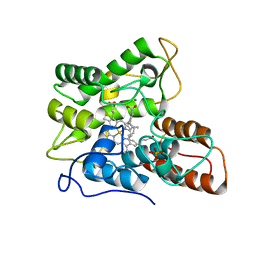
7PN7
 
 | |
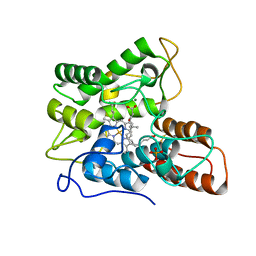
7PNA
 
 | |
7PN8
 
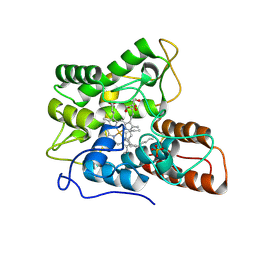 | |
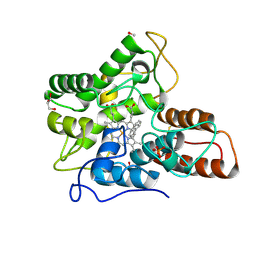
7PN4
 
 | |
7PN9
 
 | |
4AXE
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXD
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with AMPPNP | | Descriptor: | CITRIC ACID, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4EQV
 
 | |
4QAW
 
 | |
4AXF
 
 | | InsP5 2-K in complex with Ins(3,4,5,6)P4 plus AMPPNP | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, Myo inositol 3,4,5,6 tetrakisphosphate, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXC
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase apo form | | Descriptor: | GLYCEROL, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
5ANN
 
 | | Structure of fructofuranosidase from Xanthophyllomyces dendrorhous | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-FRUCTOFURANOSIDASE, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-09-07 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
2Z1S
 
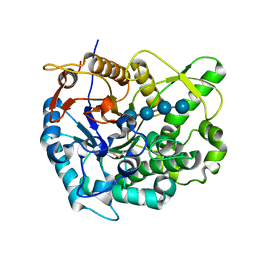 | | Beta-glucosidase B from paenibacillus polymyxa complexed with cellotetraose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Isorna, P, Sanz-Aparicio, J. | | Deposit date: | 2007-05-12 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
3LRL
 
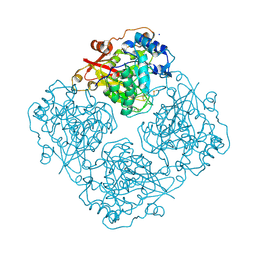 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae with melibiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
4QB6
 
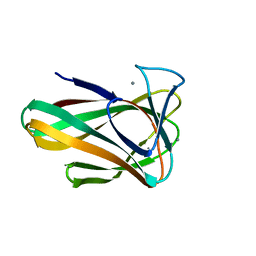 | | Structure of CBM35 in complex with aldouronic acid | | Descriptor: | CALCIUM ION, Xyn30D, alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
J.Biol.Chem., 289, 2014
|
|
4QB1
 
 | |
3LRK
 
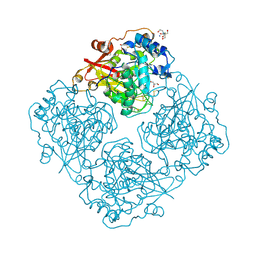 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
4QB2
 
 | | Structure of CBM35 in complex with glucuronic acid | | Descriptor: | CALCIUM ION, Xyn30D, alpha-D-glucopyranuronic acid, ... | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
J.Biol.Chem., 289, 2014
|
|
2XAL
 
 | | Lead derivative of Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAM
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAN
 
 | | inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with AMP PNP and IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MAGNESIUM ION, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
