3PMO
 
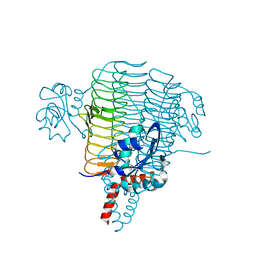 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
8T6E
 
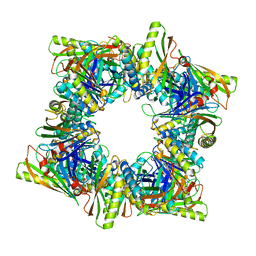 | | Crystal structure of T33-28.3: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-28.3: A, T33-28.3: B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6C
 
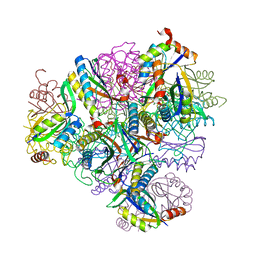 | | Crystal structure of T33-18.2: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-18.2 : A, T33-18.2 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6N
 
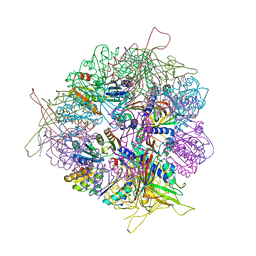 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4FS8
 
 | | The structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation | | Descriptor: | Arsenic methyltransferase, CALCIUM ION | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
9DAP
 
 | | Human norovirus GII.3 protease in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Genome polyprotein | | Authors: | Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9D9Y
 
 | | Human norovirus GI.1 Norwalk protease in complex with rupintrivir | | Descriptor: | 3C-like protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Zhao, B, Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9DF5
 
 | |
9DAL
 
 | | Human norovirus GII.3 R112A mutant protease in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Peptidase C37 | | Authors: | Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9DA0
 
 | | Human norovirus GII.4 Houston R112A mutant protease in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Peptidase C37 | | Authors: | Zhao, B, Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9DA7
 
 | | Human norovirus GII.4 Houston protease in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Peptidase C37 | | Authors: | Zhao, B, Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9DAJ
 
 | | Human norovirus GII.4 Sydney protease in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, IODIDE ION, Peptidase C37 | | Authors: | Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
9DEY
 
 | |
8TUF
 
 | |
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
3Q6L
 
 | | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution | | Descriptor: | Fatty acid-binding protein, adipocyte | | Authors: | Prasad, G.S, Carney, D, Small, V, Sankaran, B, Zwart, P.H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution
To be Published
|
|
6W30
 
 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
6O35
 
 | |
7KHY
 
 | | Crystal structure of OXA-163 K73A in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2020-10-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KHZ
 
 | | Crystal structure of OXA-163 K73A in complex with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2020-10-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
8TN0
 
 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMR
 
 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
7K2W
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
6XT4
 
 | |
