1X7V
 
 | | Crystal structure of PA3566 from Pseudomonas aeruginosa | | Descriptor: | PA3566 protein, SULFATE ION | | Authors: | Sanders, D.A, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-16 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The X-ray crystal structure of PA3566 from Pseudomonas aureginosa at 1.8 A resolution.
Proteins, 61, 2005
|
|
2Q7V
 
 | | Crystal Structure of Deinococcus Radiodurans Thioredoxin Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Sanders, D.A.R, Obiero, J, Bonderoff, S.A, Goertzen, M.M. | | Deposit date: | 2007-06-07 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thioredoxin system from Deinococcus radiodurans.
J.Bacteriol., 192, 2010
|
|
1I8T
 
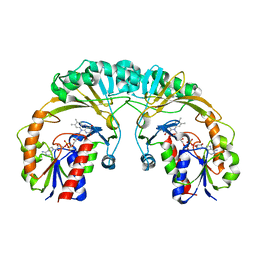 | | STRUCTURE OF UDP-GALACTOPYRANOSE MUTASE FROM E.COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Sanders, D.A.R, Staines, A.G, McMahon, S.A, McNeil, M.R, Whitfield, C, Naismith, J.H. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UDP-galactopyranose mutase has a novel structure and mechanism.
Nat.Struct.Biol., 8, 2001
|
|
1I3H
 
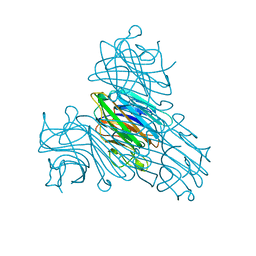 | | CONCANAVALIN A-DIMANNOSE STRUCTURE | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Sanders, D.A.R, Moothoo, D.N, Raftery, J, Howard, A.J, Helliwell, J.R, Naismith, J.H. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution structure of the Con A-dimannose complex.
J.Mol.Biol., 310, 2001
|
|
3NT5
 
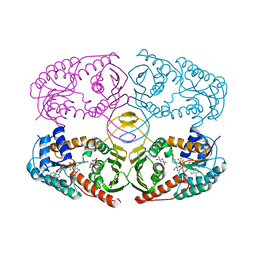 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9006 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTO
 
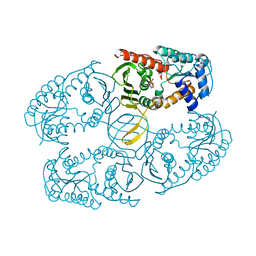 | |
3NT2
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3003 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
1U6Z
 
 | |
1X9J
 
 | |
3E4D
 
 | | Structural and Kinetic Study of an S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, Esterase D, MAGNESIUM ION | | Authors: | Van Straaten, K.E, Gonzalez, C.F, Valladares, R.B, Xu, X, Savchenko, A.V, Sanders, D.A.R. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
Protein Sci., 18, 2009
|
|
4RPJ
 
 | |
4RPK
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galf | | Descriptor: | (2R,5S)-5-[(1R)-1,2-dihydroxyethyl]-3,3,4,4-tetrafluorotetrahydrofuran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPH
 
 | |
4RPG
 
 | |
4RPL
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
3UKL
 
 | |
3UKK
 
 | |
3UKQ
 
 | |
3UKA
 
 | |
3UKP
 
 | |
3UKH
 
 | |
3UKF
 
 | |
3NTR
 
 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6503 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NT4
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTQ
 
 | |
