4MO2
 
 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZD
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the reduced internal aldimine and with bound Glutarate | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
4MIO
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD(H) and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD(H) and myo-inositol
To be Published
|
|
4MIE
 
 | |
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
6V2N
 
 | | Crystal structure of E. coli phosphoenolpyruvate carboxykinase mutant Lys254Ser | | Descriptor: | ACETATE ION, CALCIUM ION, Phosphoenolpyruvate carboxykinase (ATP) | | Authors: | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6V2L
 
 | | E. coli Phosphoenolpyruvate carboxykinase S250A | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Sokaribo, A.S, Goldie, H, Sanders, D. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
4MJL
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol | | Descriptor: | (1R,2R,3S,4S,5S,6S)-CYCLOHEXANE-1,2,3,4,5,6-HEXOL, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-03 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol
To be Published
|
|
4MIY
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol
To be Published
|
|
7KEL
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To Be Published
|
|
4N54
 
 | | Crystal structure of scyllo-inositol dehydrogenase from Lactobacillus casei with bound cofactor NAD(H) and scyllo-inositol | | Descriptor: | (1r,2r,3r,4r,5r,6r)-cyclohexane-1,2,3,4,5,6-hexol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol dehydrogenase | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-10-09 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of scyllo-inositol dehydrogenase from Lactobacillus casei with bound cofactor NAD(H) and scyllo-inositol
To be Published
|
|
4MIN
 
 | |
4MKZ
 
 | |
4MKX
 
 | |
1G99
 
 | | AN ANCIENT ENZYME: ACETATE KINASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | ACETATE KINASE, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Buss, K.A, Cooper, D.R, Ingram-Smith, C, Ferry, J.G, Sanders, D.A, Hasson, M.S. | | Deposit date: | 2000-11-22 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases.
J.Bacteriol., 183, 2001
|
|
1SAZ
 
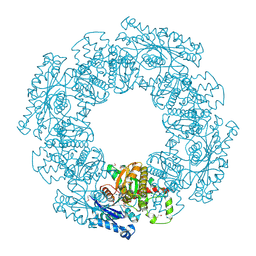 | | Membership in the ASKHA Superfamily: Enzymological Properties and Crystal Structure of Butyrate Kinase 2 from Thermotoga maritima | | Descriptor: | FORMIC ACID, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable butyrate kinase 2, ... | | Authors: | Diao, J, Cooper, D.R, Sanders, D.A, Hasson, M.S. | | Deposit date: | 2004-02-09 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of butyrate kinase 2 from Thermotoga maritima, a member of the ASKHA superfamily of phosphotransferases.
J.Bacteriol., 191, 2009
|
|
7KUZ
 
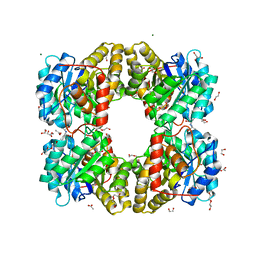 | |
7KKD
 
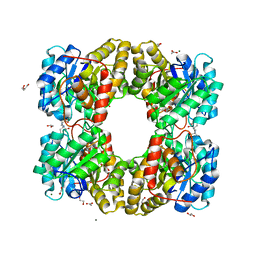 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
7KOC
 
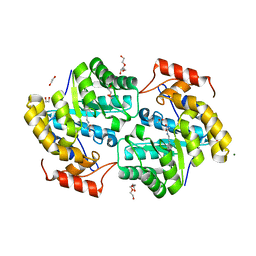 | |
7KPC
 
 | |
7KR7
 
 | |
