4RCZ
 
 | | Structure of aIF2-gamma D19A variant from Sulfolobus solfataricus bound to GDPNP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
4RD1
 
 | | Structure of aIF2-gamma H97A variant from Sulfolobus solfataricus bound to GTP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
4RCY
 
 | | Structure of aIF2-gamma D19A variant from Sulfolobus solfataricus bound to GTP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
4RD0
 
 | | Structure of aIF2-gamma D19A variant from Sulfolobus solfataricus bound to GDP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
3PGF
 
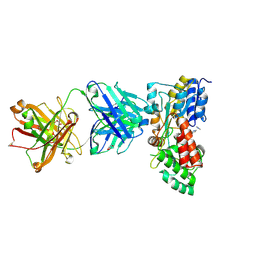 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7PFM
 
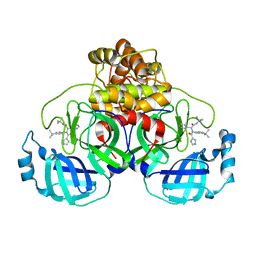 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
3GD8
 
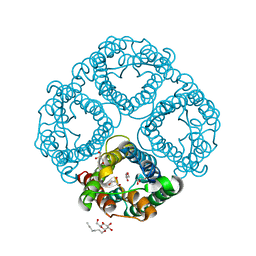 | | Crystal Structure of Human Aquaporin 4 at 1.8 and its Mechanism of Conductance | | Descriptor: | Aquaporin-4, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Ho, J.D, Yeh, R, Sandstrom, A, Chorny, I, Harries, W.E.C, Robbins, R.A, Miercke, L.J.W, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1GR7
 
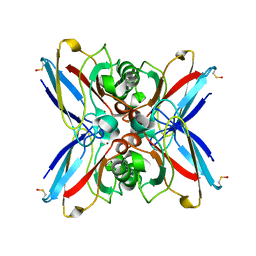 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
