6QS7
 
 | |
6QS6
 
 | |
4D2U
 
 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2Q
 
 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
8BA7
 
 | |
8BA8
 
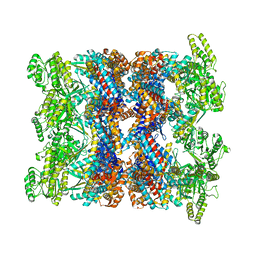 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BA9
 
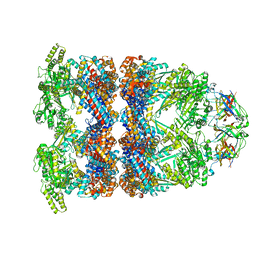 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8B6U
 
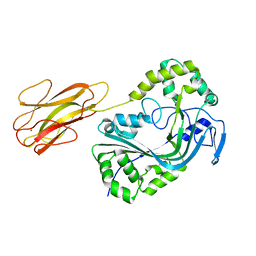 | | Mpf2Ba1 monomer | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez-Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6W
 
 | | Mpf2Ba1 pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6V
 
 | | Mp2Ba1 pre-pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8A00
 
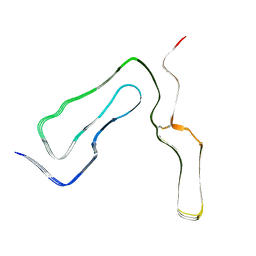 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2BYU
 
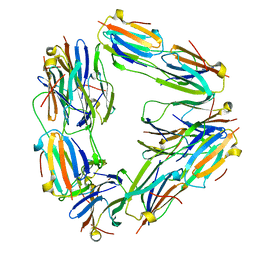 | | Negative stain EM reconstruction of M.tuberculosis Acr1(Hsp 16.3) fitted with wheat sHSP dimer | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Kennaway, C.K, Benesch, J.L.P, Gohlke, U, Wang, L, Robinson, C.V, Orlova, E.V, Saibil, H.R, Keep, N.H. | | Deposit date: | 2005-08-05 | | Release date: | 2005-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dodecameric Structure of the Small Heat Shock Protein Acr1 from Mycobacterium Tuberculosis.
J.Biol.Chem., 280, 2005
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2CGT
 
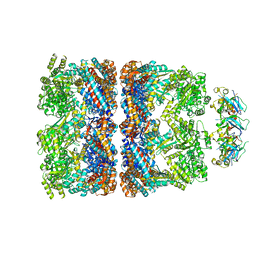 | | GROEL-ADP-gp31 COMPLEX | | Descriptor: | 60 KDA GROEL, CAPSID ASSEMBLY PROTEIN GP31 | | Authors: | Clare, D.K, Bakkes, P.J, van Heerikhuizen, H, van der Vies, S.M, Saibil, H.R. | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | An Expanded Protein Folding Cage in the Groel-Gp31 Complex.
J.Mol.Biol., 358, 2006
|
|
2C7E
 
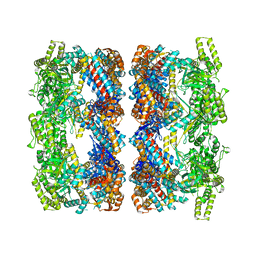 | | REVISED ATOMIC STRUCTURE FITTING INTO A GROEL(D398A)-ATP7 CRYO-EM MAP (EMD 1047) | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy
Cell(Cambridge,Mass.), 107, 2001
|
|
2C7D
 
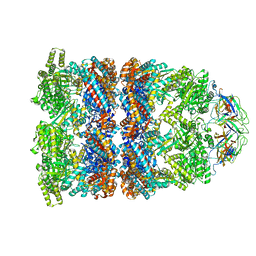 | | Fitted coordinates for GroEL-ADP7-GroES Cryo-EM complex (EMD-1181) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2C7C
 
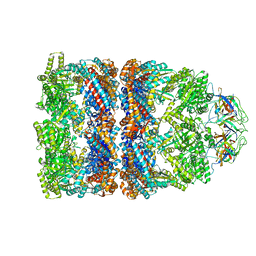 | | FITTED COORDINATES FOR GROEL-ATP7-GROES CRYO-EM COMPLEX (EMD-1180) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4A8D
 
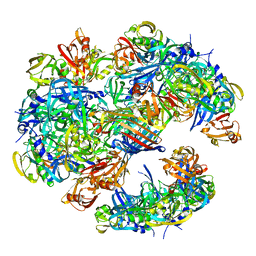 | | DegP dodecamer with bound OMP | | Descriptor: | OUTER MEMBRANE PROTEIN C, PERIPLASMIC SERINE ENDOPROTEASE DEGP | | Authors: | Malet, H, Krojer, T, Sawa, J, Schafer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A9G
 
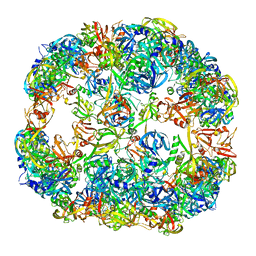 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AAR
 
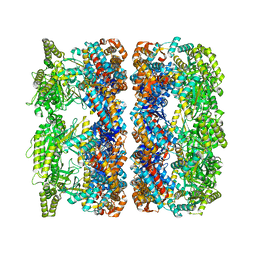 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4A8C
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AAS
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
