6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MGK
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
2PC6
 
 | | Crystal structure of putative acetolactate synthase- small subunit from Nitrosomonas europaea | | Descriptor: | CALCIUM ION, Probable acetolactate synthase isozyme III (Small subunit), UNKNOWN LIGAND | | Authors: | Petkowski, J.J, Chruszcz, M, Zimmerman, M.D, Zheng, H, Cymborowski, M.T, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of TM0549 and NE1324--two orthologs of E. coli AHAS isozyme III small regulatory subunit.
Protein Sci., 16, 2007
|
|
2O30
 
 | | Nuclear movement protein from E. cuniculi GB-M1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NUCLEAR MOVEMENT PROTEIN | | Authors: | Binkowski, T.A, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Nuclear movement protein from E. cuniculi GB-M1
TO BE PUBLISHED
|
|
3OS6
 
 | | Crystal structure of putative 2,3-dihydroxybenzoate-specific isochorismate synthase, DhbC from Bacillus anthracis. | | Descriptor: | GLYCEROL, Isochorismate synthase DhbC, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Domagalski, M.J, Chruszcz, M, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of isochorismate synthase DhbC from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2FBL
 
 | | The crystal structure of the hypothetical protein NE1496 | | Descriptor: | SODIUM ION, hypothetical protein NE1496 | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the hypothetical protein NE1496
To be Published
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
1JQ3
 
 | | Crystal Structure of Spermidine Synthase in Complex with Transition State Analogue AdoDATO | | Descriptor: | S-ADENOSYL-1,8-DIAMINO-3-THIOOCTANE, Spermidine synthase | | Authors: | Korolev, S, Ikeguchi, Y, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Pegg, A.E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
1K4N
 
 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1KS2
 
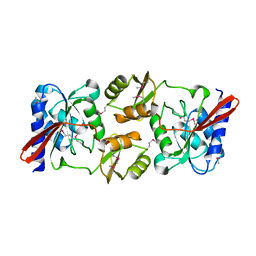 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
1KXJ
 
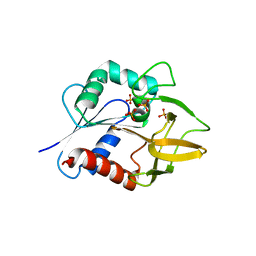 | | The Crystal Structure of Glutamine Amidotransferase from Thermotoga maritima | | Descriptor: | Amidotransferase hisH, PHOSPHATE ION | | Authors: | Korolev, S, Skarina, T, Evdokimova, E, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Thermotoga maritima
Proteins, 49, 2002
|
|
1KTN
 
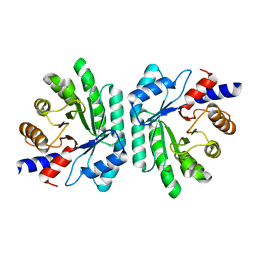 | | Structural Genomics, Protein EC1535 | | Descriptor: | 2-deoxyribose-5-phosphate aldolase | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Skarina, T, Evdokimova, E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-16 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.5A crystal structure of
2-deoxyribose-5-phosphate aldlase
To be Published
|
|
1PW5
 
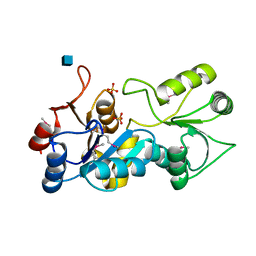 | | putative nagD protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, SULFATE ION, nagD protein, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-30 | | Release date: | 2004-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | putative nagD protein
To be Published
|
|
5T1P
 
 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
1INL
 
 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
2FXA
 
 | | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis. | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis.
TO BE PUBLISHED
|
|
2IA2
 
 | | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1 | | Descriptor: | Putative Transcriptional Regulator | | Authors: | Cymborowski, M.T, Chruszcz, M, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1
To be Published
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | Descriptor: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7TOK
 
 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | Descriptor: | Acetylxylan esterase I | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOG
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | Descriptor: | SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
