1YW0
 
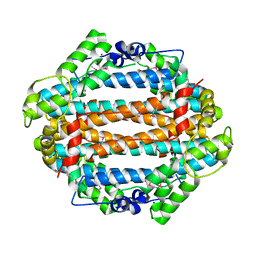 | | Crystal structure of the tryptophan 2,3-dioxygenase from Xanthomonas campestris. Northeast Structural Genomics Target XcR13. | | Descriptor: | MAGNESIUM ION, tryptophan 2,3-dioxygenase | | Authors: | Vorobiev, S.M, Abashidze, M, Forouhar, F, Kuzin, A, Xiao, R, Ciano, M, Aton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tryptophan 2,3-dioxygenase from Xanthomonas campestris. Northeast Structural Genomics Target XcR13.
To be Published
|
|
242D
 
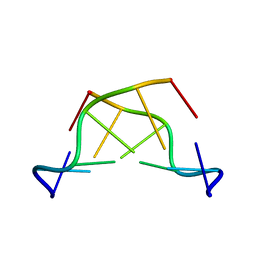 | | MAD PHASING STRATEGIES EXPLORED WITH A BROMINATED OLIGONUCLEOTIDE CRYSTAL AT 1.65 A RESOLUTION. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(CBR)P*G)-3') | | Authors: | Peterson, M.R, Harrop, S.J, McSweeney, S.M, Leonard, G.A, Thompson, A.W, Hunter, W.N, Helliwell, J.R. | | Deposit date: | 1996-06-20 | | Release date: | 1996-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MAD Phasing Strategies Explored with a Brominated Oligonucleotide Crystal at 1.65A Resolution.
J.Synchrotron Radiat., 3, 1996
|
|
2CG9
 
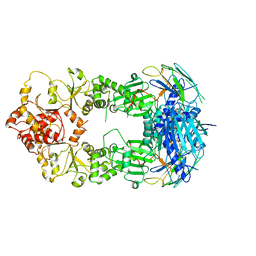 | | Crystal structure of an Hsp90-Sba1 closed chaperone complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CO-CHAPERONE PROTEIN SBA1 | | Authors: | Ali, M.M.U, Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex
Nature, 440, 2006
|
|
2CGE
 
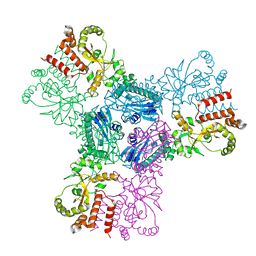 | |
2C7Y
 
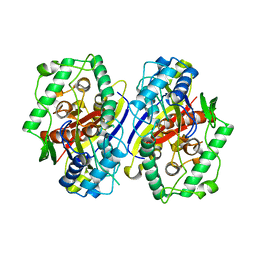 | | plant enzyme | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Germain, V, Bryce, J.H, Smith, S.M, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
2B70
 
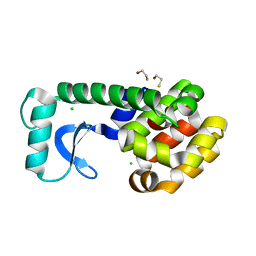 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B75
 
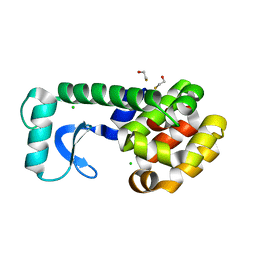 | | T4 Lysozyme mutant L99A at 150 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6W
 
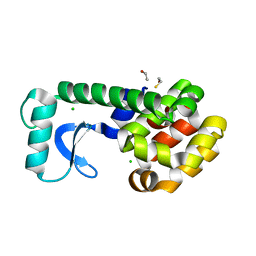 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B72
 
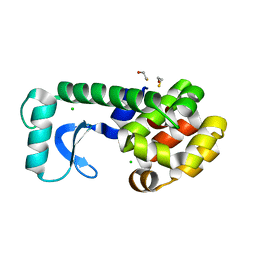 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6Z
 
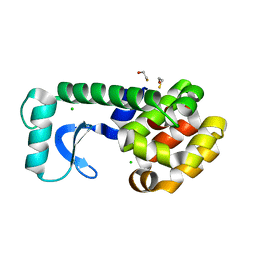 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6X
 
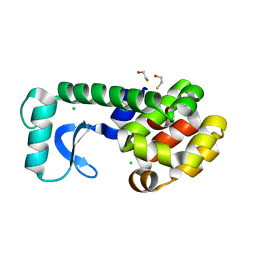 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2CFK
 
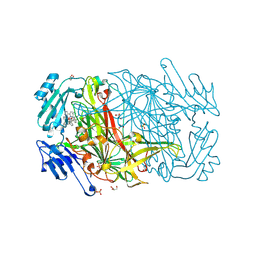 | | AGAO in complex with wc5 (Ru-wire inhibitor, 5-carbon linker) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-21 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enantiomer-specific binding of ruthenium(II) molecular wires by the amine oxidase of Arthrobacter globiformis.
J. Am. Chem. Soc., 130, 2008
|
|
2CM4
 
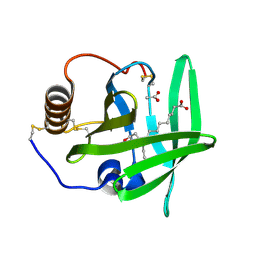 | | The complement inhibitor OmCI in complex with ricinoleic acid | | Descriptor: | ACETATE ION, COMPLEMENT INHIBITOR, RICINOLEIC ACID | | Authors: | Roversi, P, Johnson, S, Lissina, O, Paesen, G.C, Boland, W, Nunn, M.A, Lea, S.M. | | Deposit date: | 2006-05-04 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Omci, a Novel Lipocalin Inhibitor of the Complement System.
J.Mol.Biol., 369, 2007
|
|
2CA9
 
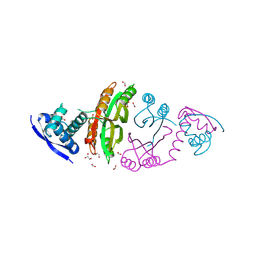 | | apo-NIKR from helicobacter pylori in closed trans-conformation | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CAD
 
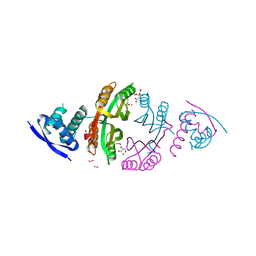 | | NikR from Helicobacter pylori in closed trans-conformation and nickel bound to 2F, 2X and 2I sites. | | Descriptor: | CITRIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CJD
 
 | |
2CJG
 
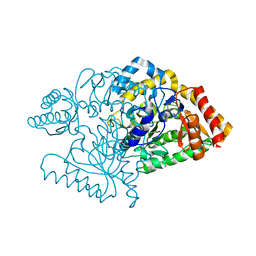 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2CTN
 
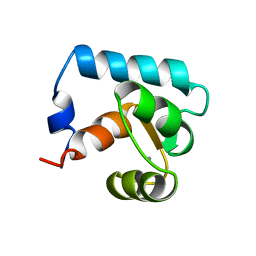 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
2D3O
 
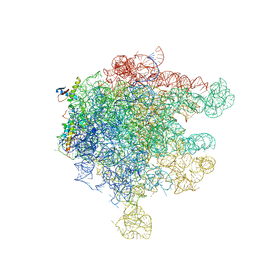 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
7ON8
 
 | |
7ONG
 
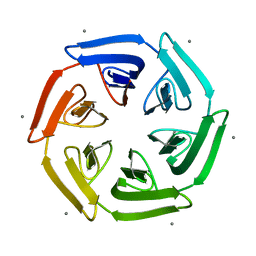 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | Descriptor: | CALCIUM ION, SAKe6BE-L1 | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONA
 
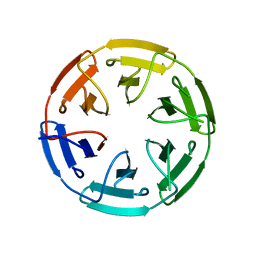 | | Crystal structure of the computationally designed SAKe6AC protein | | Descriptor: | CALCIUM ION, SAKe6AC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OPU
 
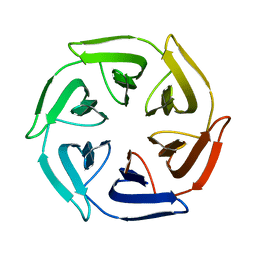 | |
7ON7
 
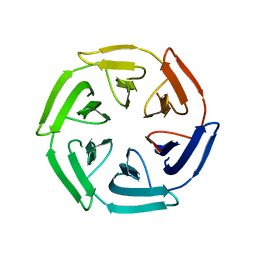 | |
7ONE
 
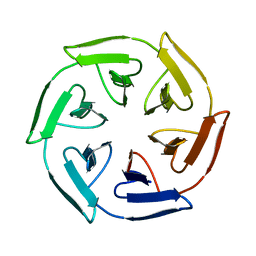 | | Crystal structure of the self-assembled SAKe6BE designer protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
