8SA6
 
 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
1IL9
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 8-METHYL-9-OXOGUANINE | | Descriptor: | 5-AMINO-2-METHYL-6H-OXAZOLO[5,4-D]PYRIMIDIN-7-ONE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
5IOT
 
 | |
2XL3
 
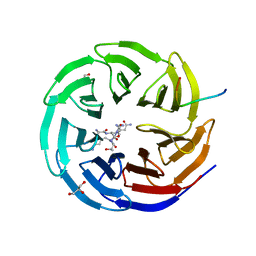 | | WDR5 IN COMPLEX WITH AN RBBP5 PEPTIDE AND HISTONE H3 PEPTIDE | | Descriptor: | GLYCEROL, HISTONE H3.1, RETINOBLASTOMA-BINDING PROTEIN 5, ... | | Authors: | Odho, Z, Southall, S.M, Wilson, J.R. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterisation of a Novel Wdr5 Binding Site that Recruits Rbbp5 Through a Conserved Motif and Enhances Methylation of H3K4 by Mll1.
J.Biol.Chem., 285, 2010
|
|
2XFD
 
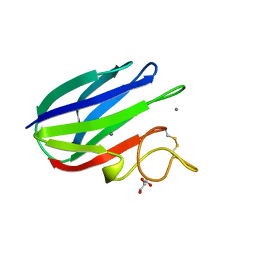 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
1I6T
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Samygina, V.R, Popov, A.N, Lamzin, V.S, Avaeva, S.M. | | Deposit date: | 2001-03-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structures of Escherichia coli inorganic pyrophosphatase complexed with Ca(2+) or CaPP(i) at atomic resolution and their mechanistic implications.
J.Mol.Biol., 314, 2001
|
|
7KJG
 
 | | F96M epi-isozizaene synthase: complex with 3 Mg2+ and BTAC | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
7KJD
 
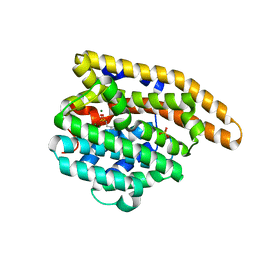 | | F96M epi-isozizaene synthase: complex with 3 Mg2+ and risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
2XRC
 
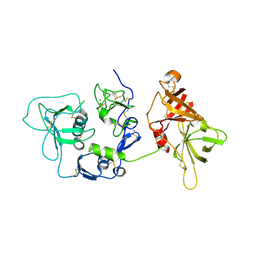 | | Human complement factor I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HUMAN COMPLEMENT FACTOR I | | Authors: | Roversi, P, Johnson, S, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Basis for Complement Factor I Control and its Disease-Associated Sequence Polymorphisms.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XX5
 
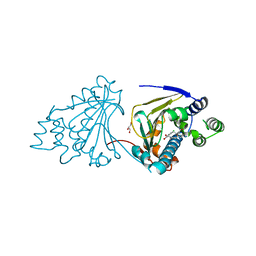 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E,10R)-N-BENZYL-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDRO-2-BENZAZACYCLOTETRADECINE-10-CARBOXAMIDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2XZ7
 
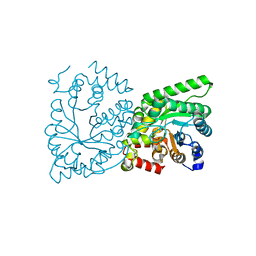 | | CRYSTAL STRUCTURE OF THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PHOSPHOENOLPYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA) | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
5H6H
 
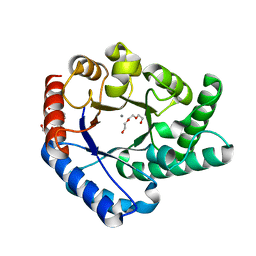 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ribulose 3-Epimerase with Mn2+ | | Descriptor: | MANGANESE (II) ION, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Cao, T.-P, Choi, J.M, Shin, S.M, Le, D.W, Lee, S.H. | | Deposit date: | 2016-11-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
2XXS
 
 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
2VJ5
 
 | | Shigella flexneri MxiC | | Descriptor: | PROTEIN MXIC | | Authors: | Deane, J.E, Roversi, P, King, C, Johnson, S, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Shigella Flexneri Type 3 Secretion System Protein Mxic Reveal Conformational Variability Amongst Homologues.
J.Mol.Biol., 377, 2008
|
|
7SNL
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide
To Be Published
|
|
2XZ9
 
 | | CRYSTAL STRUCTURE FROM THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA), PYRUVIC ACID | | Authors: | Oberholzer, A.E, Waltersperger, S.M, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
7SNN
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide
To Be Published
|
|
1IZ3
 
 | | Dimeric structure of FIH (Factor inhibiting HIF) | | Descriptor: | FIH, SULFATE ION | | Authors: | Lee, C, Kim, S.-J, Jeong, D.-G, Lee, S.M, Ryu, S.-E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
J.Biol.Chem., 278, 2003
|
|
7UY4
 
 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
8T94
 
 | |
2XRD
 
 | | Structure of the N-terminal four domains of the complement regulator Rat Crry | | Descriptor: | COMPLEMENT REGULATORY PROTEIN CRRY | | Authors: | Leath, K.J, Roversi, P, Johnson, S, Morgan, B.P, Lea, S.M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of the Rat Complement Regulator Crry.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
7KJF
 
 | | Wild-type epi-isozizaene synthase: complex with 3 Mg2+ and neridronate | | Descriptor: | (6-azanyl-1-oxidanyl-1-phosphono-hexyl)phosphonic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
