2OE7
 
 | | High-Pressure T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
2OEA
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
1TTZ
 
 | | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Ho, C.K, Cooper, B, Ma, L.-C, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris
To be Published
|
|
6B25
 
 | |
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
3EW7
 
 | | Crystal structure of the Lmo0794 protein from Listeria monocytogenes. Northeast Structural Genomics Consortium target LmR162. | | Descriptor: | Lmo0794 protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the Lmo0794 protein from Listeria monocytogenes.
To be Published
|
|
4XE6
 
 | |
6U7J
 
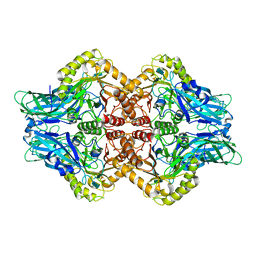 | | Uncultured Clostridium sp. Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CALCIUM ION | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens.
J.Biol.Chem., 294, 2019
|
|
6B29
 
 | |
1K8J
 
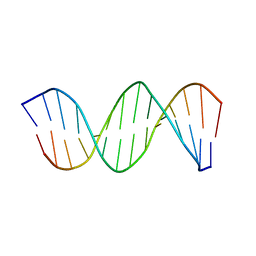 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
4LOA
 
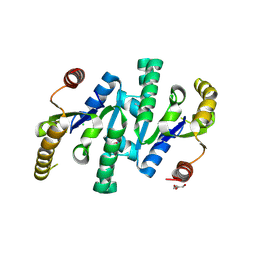 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
1KJ9
 
 | | Crystal structure of purt-encoded glycinamide ribonucleotide transformylase complexed with Mg-ATP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
3KBG
 
 | | Crystal structure of the 30S ribosomal protein S4e from Thermoplasma acidophilum. Northeast Structural Genomics Consortium Target TaR28. | | Descriptor: | 30S ribosomal protein S4e | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Belote, R.L, Ciccosanti, C, Patel, D, Liu, J, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the 30S ribosomal protein S4e from Thermoplasma acidophilum.
To be Published
|
|
1TNQ
 
 | |
3FGB
 
 | | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR289b. | | Descriptor: | uncharacterized protein Q89ZH8_BACTN | | Authors: | Vorobiev, S.M, Lew, S, Seetharaman, J, Lee, D, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron.
To be Published
|
|
1K86
 
 | | Crystal structure of caspase-7 | | Descriptor: | caspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
1TO0
 
 | | X-ray structure of Northeast Structural Genomics target protein sr145 from Bacillus subtilis | | Descriptor: | Hypothetical UPF0247 protein yyda | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Shastry, R, Ma, L.-C, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein sr145 from Bacillus subtilis
To be Published
|
|
5CD4
 
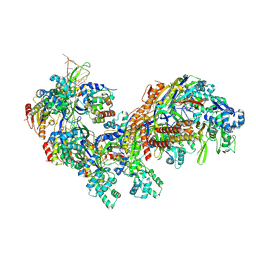 | | The Type IE CRISPR Cascade complex from E. coli, with two assemblies in the asymmetric unit arranged back-to-back | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2015-07-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
3FIA
 
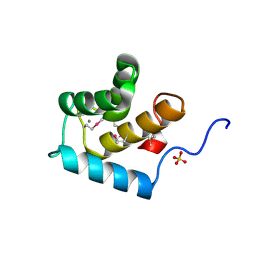 | | Crystal structure of the EH 1 domain from human intersectin-1 protein. Northeast Structural Genomics Consortium target HR3646e. | | Descriptor: | CALCIUM ION, Intersectin-1, SULFATE ION | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Devices, C, Zhao, L, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the EH 1 domain from human intersectin-1 protein.
To be Published
|
|
4XHO
 
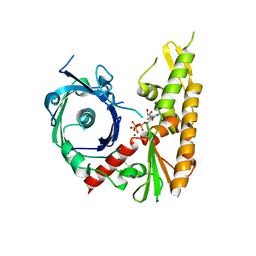 | | Bacillus thuringiensis ParM with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4LTM
 
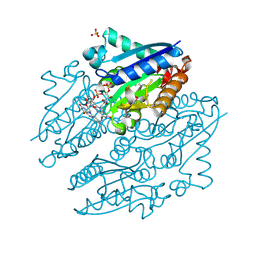 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
8EHG
 
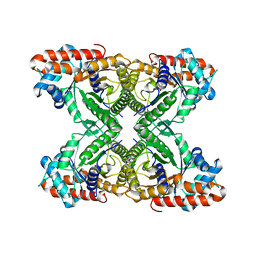 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
1TZA
 
 | | X-ray structure of Northeast Structural Genomics Consortium target SoR45 | | Descriptor: | CACODYLATE ION, apaG protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Edstrom, W, Acton, T.B, Shastry, R, Ma, L.-C, Cooper, B, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of Northeast Structural Genomics Consortium
target SoR45
To be Published
|
|
