5B7Y
 
 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Co2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COBALT (II) ION, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
1ODG
 
 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
1BA0
 
 | | HEAT-SHOCK COGNATE 70KD PROTEIN 44KD ATPASE N-TERMINAL 1NGE 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HEAT-SHOCK COGNATE 70KD PROTEIN, ... | | Authors: | Wilbanks, S.M, Mckay, D.B. | | Deposit date: | 1998-04-21 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural replacement of active site monovalent cations by the epsilon-amino group of lysine in the ATPase fragment of bovine Hsc70.
Biochemistry, 37, 1998
|
|
3N0L
 
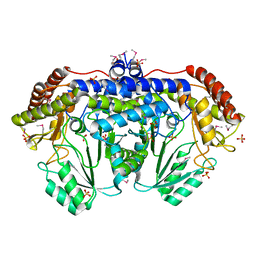 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
4OSF
 
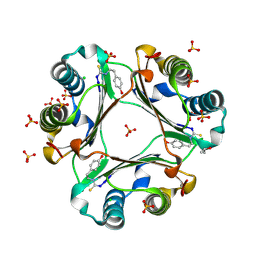 | | 4-(2-isothiocyanatoethyl)phenol inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
4OYQ
 
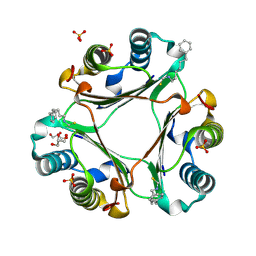 | | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | 6-isothiocyanatohexylbenzene, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | To be published
To be published
|
|
1NMD
 
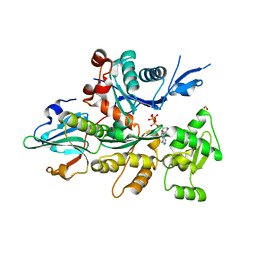 | | Crystal Structure of D. Discoideum Actin-Gelsolin Segment 1 Complex Crystallized In Presence Of Lithium ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Vorobiev, S.M, Welti, S, Condeelis, J, Almo, S.C. | | Deposit date: | 2003-01-09 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure Of The Non-Vertebrate Actin: Implications For The ATP Hydrolytic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1BGQ
 
 | |
5AZJ
 
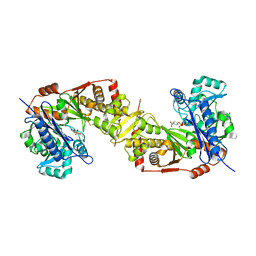 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with 4NP (with disulfide bridge) | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tokuoka, S.M, Tokumasu, F, Sakamoto, K, Michels, P.A.M, Harada, S, Kita, K. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Glycerol kinase of African trypanosomes possesses an intrinsic phosphatase activity.
Biochim Biophys Acta Gen Subj, 1861, 2017
|
|
1O9U
 
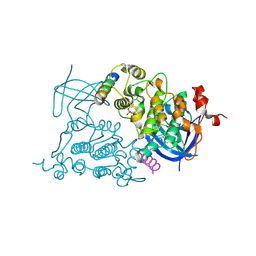 | |
1O7U
 
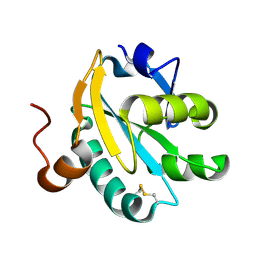 | | Radiation induced tryparedoxin-I | | Descriptor: | TRYPAREDOXIN | | Authors: | Alphey, M.S, Bond, C.S, McSweeney, S.M, Hunter, W.N. | | Deposit date: | 2002-11-14 | | Release date: | 2003-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
3HFQ
 
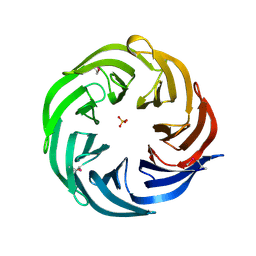 | | Crystal structure of the lp_2219 protein from Lactobacillus plantarum. Northeast Structural Genomics Consortium Target LpR118. | | Descriptor: | PHOSPHATE ION, uncharacterized protein lp_2219 | | Authors: | Vorobiev, S.M, Scott, L, Schauder, C, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Crystal structure of the lp_2219 protein from Lactobacillus plantarum.
To be Published
|
|
4PHY
 
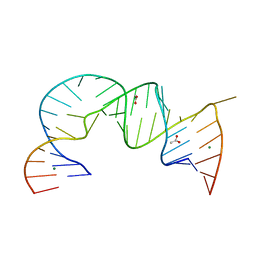 | | Functional conservation despite structural divergence in ligand-responsive RNA switches | | Descriptor: | ACETATE ION, MAGNESIUM ION, RNA (26-MER), ... | | Authors: | Boerneke, M.A, Dibrov, S.M, Hermann, T.H. | | Deposit date: | 2014-05-07 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional conservation despite structural divergence in ligand-responsive RNA switches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3F95
 
 | | Crystal Structure of Extra C-terminal Domain (X) of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | Descriptor: | Beta-glucosidase, CHLORIDE ION | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2008-11-13 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
3N2Z
 
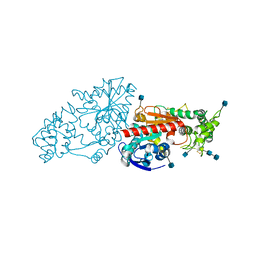 | | The Structure of Human Prolylcarboxypeptidase at 2.80 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal Pro-X carboxypeptidase, ... | | Authors: | Soisson, S.M, Patel, S.B, Lumb, K.J, Sharma, S. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural definition and substrate specificity of the S28 protease family: the crystal structure of human prolylcarboxypeptidase.
Bmc Struct.Biol., 10, 2010
|
|
3F1T
 
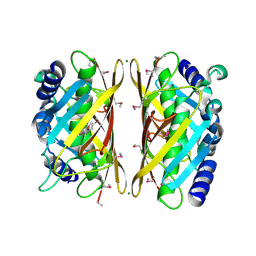 | | Crystal structure of the Q9I3C8_PSEAE protein from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR319a. | | Descriptor: | MAGNESIUM ION, uncharacterized protein Q9I3C8_PSEAE | | Authors: | Vorobiev, S.M, Chen, Y, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Q9I3C8_PSEAE protein from Pseudomonas aeruginosa.
To be Published
|
|
4Q90
 
 | |
4PZO
 
 | | Crystal structure of PHC3 SAM L967R | | Descriptor: | Polyhomeotic-like protein 3 | | Authors: | Nanyes, D.R, Junco, S.E, Taylor, A.B, Robinson, A.K, Patterson, N.L, Shivarajpur, A, Halloran, J, Hale, S.M, Kaur, Y, Hart, P.J, Kim, C.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Proteins, 82, 2014
|
|
4Q7P
 
 | |
4Q87
 
 | |
4Q8Y
 
 | |
1NVS
 
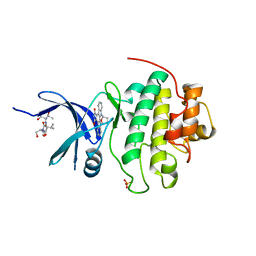 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
3GOZ
 
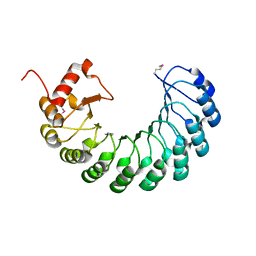 | | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148 | | Descriptor: | Leucine-rich repeat-containing protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Belote, R.L, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-20 | | Release date: | 2009-03-31 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148.
To be Published
|
|
4MN4
 
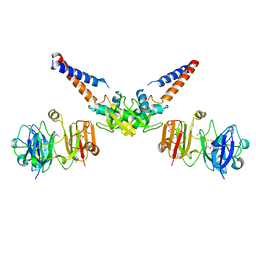 | | Structural Basis for the MukB-topoisomerase IV Interaction | | Descriptor: | Chromosome partition protein MukB, DNA topoisomerase 4 subunit A | | Authors: | Vos, S.M, Stewart, N.K, Oakley, M.G, Berger, J.M. | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.
Embo J., 32, 2013
|
|
