5EJX
 
 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2GZX
 
 | | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237. | | Descriptor: | NICKEL (II) ION, putative TatD related DNase | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Wang, D, Fang, Y, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237.
To be Published
|
|
2BOU
 
 | | EGF Domains 1,2,5 of human EMR2, a 7-TM immune system molecule, in complex with barium. | | Descriptor: | BARIUM ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Abbott, R.J.M, Spendlove, I, Roversi, P, Teriete, P, Knott, V, Handford, P.A, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2005-04-14 | | Release date: | 2006-10-18 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of a Novel T Cell Receptor Co-Regulatory Protein Complex, Cd97-Cd55.
J.Biol.Chem., 282, 2007
|
|
1DQD
 
 | | CRYSTAL STRUCTURE OF FAB HGR-2 F6, A COMPETITIVE ANTAGONIST OF THE GLUCAGON RECEPTOR | | Descriptor: | FAB HGR-2 F6 | | Authors: | Wright, L.M, Brzozowski, A.M, Hubbard, R.E, Pike, A.C.W, Roberts, S.M, Skovgaard, R.N, Svendsen, I, Vissing, H, Bywater, R.P. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Fab hGR-2 F6, a competitive antagonist of the glucagon receptor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2BG8
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT and 1mM TCEP-HCl were used as reducing agents. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O24
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
5EXQ
 
 | | Human cytochrome c Y48H | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Fellner, M, Jameson, G.N.L, Ledgerwood, E.C, Wilbanks, S.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity.
Biochem.J., 2021
|
|
2HLQ
 
 | |
5EZW
 
 | |
2BGA
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH7 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2HLR
 
 | |
4NZV
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
2C8I
 
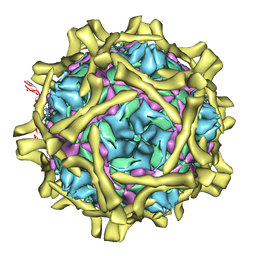 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
2CA5
 
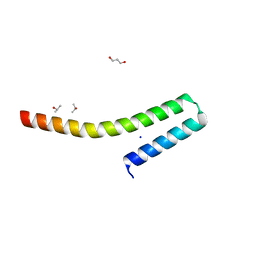 | | MxiH needle protein of Shigella Flexneri (monomeric form, residues 1- 78) | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, MXIH, ... | | Authors: | Deane, J.E, Roversi, P, Cordes, F.S, Johnson, S, Kenjale, R, Picking, W.L, Picking, W.D, Blocker, A.J, Lea, S.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Model of a Type III Secretion System Needle: Implications for Host-Cell Sensing
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1ZAC
 
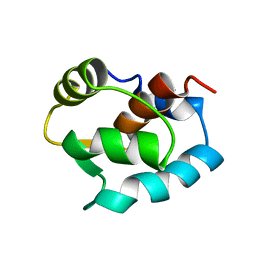 | | N-DOMAIN OF TROPONIN C FROM CHICKEN SKELETAL MUSCLE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
4B4A
 
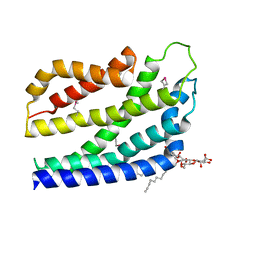 | | Structure of the TatC core of the twin arginine protein translocation system | | Descriptor: | Lauryl Maltose Neopentyl Glycol, SEC-INDEPENDENT PROTEIN TRANSLOCASE PROTEIN TATC | | Authors: | Rollauer, S.E, Tarry, M.J, Jaaskelainen, M, Graham, J.E, Jaeger, F, Krehenbrink, M, Roversi, P, McDowell, M.A, Stansfeld, P.J, Johnson, S, Liu, S.M, Lukey, M.J, Marcoux, J, Robinson, C.V, Sansom, M.S, Palmer, T, Hogbom, M, Berks, B.C, Lea, S.M. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-05 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Nature, 49, 2012
|
|
5EE7
 
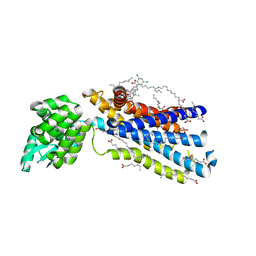 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
1F37
 
 | | STRUCTURE OF A THIOREDOXIN-LIKE [2FE-2S] FERREDOXIN FROM AQUIFEX AEOLICUS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S], GLYCEROL | | Authors: | Yeh, A.P, Chatelet, C, Soltis, S.M, Kuhn, P, Meyer, J, Rees, D.C. | | Deposit date: | 2000-05-31 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus.
J.Mol.Biol., 300, 2000
|
|
5EFU
 
 | |
2HF1
 
 | | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium violaceum. NESG target CvR39. | | Descriptor: | Tetraacyldisaccharide-1-P 4-kinase, ZINC ION | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Chen, C.X, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium
violaceum.
To be Published
|
|
4O9W
 
 | |
2HAD
 
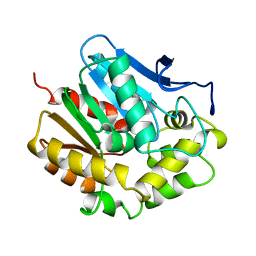 | |
1C4B
 
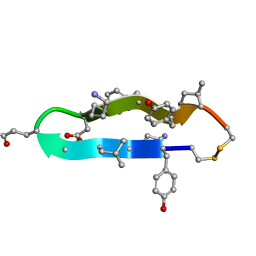 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
2HQV
 
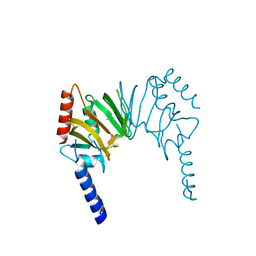 | | X-ray Crystal Structure of Protein AGR_C_4470 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR92. | | Descriptor: | AGR_C_4470p | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Zhao, L, Cunningham, K, Ma, L.C, Fang, Y, Xiao, R, Acton, T, Montelione, T.G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AGR_C_4470p from Agrobacterium tumefaciens.
Protein Sci., 16, 2007
|
|
