2QGO
 
 | |
2QYZ
 
 | | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88 | | Descriptor: | Uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Wasserman, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-15 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88.
To be Published
|
|
2WLS
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with AMTS13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, Y.P, Ekstrom, F, Polsinelli, G.A, Gao, Y, Rana, S, Hua, D.H, Andersson, B, Andersson, P.O, Peng, L, Singh, S.K, Mishra, R.K, Zhu, K.Y, Fallon, A.M, Ragsdale, D.W, Brimijoin, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective and Irreversible Inhibitors of Mosquito Acetylcholinesterases for Controlling Malaria and Other Mosquito-Borne Diseases.
Plos One, 4, 2009
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
4HCQ
 
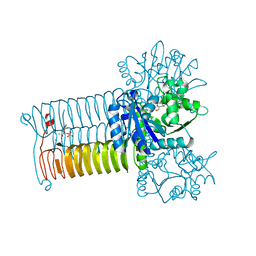 | | Crystal structure of GLMU from mycobacterium tuberculosis in complex with glucosamine-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, COBALT (II) ION, ... | | Authors: | Jagtap, P.K.A, Verma, S.K, Vithani, N. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures identify an atypical two-metal-ion mechanism for uridyltransfer in GlmU: its significance to sugar nucleotidyl transferases
J.Mol.Biol., 425, 2013
|
|
4GS1
 
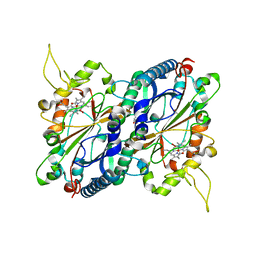 | | Crystal structure of DyP-type peroxidase from Thermobifida cellulosilytica | | Descriptor: | DyP-type peroxidase, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4H6W
 
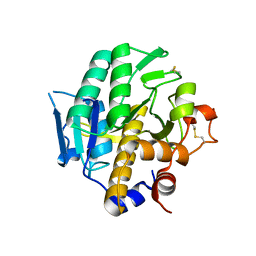 | |
4GYP
 
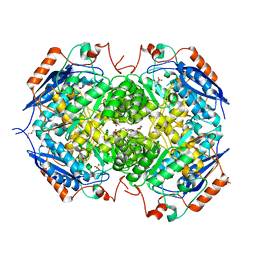 | | Crystal structure of the heterotetrameric complex of GlucD and GlucDRP from E. coli K-12 MG1655 (EFI TARGET EFI-506058) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
4H6X
 
 | | Structure of Patellamide maturation protease PatG | | Descriptor: | Thiazoline oxidase/subtilisin-like protease | | Authors: | Nair, S.K, Agarwal, V. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cyanobactin maturation enzymes define a family of transamidating proteases.
Chem.Biol., 19, 2012
|
|
4HL7
 
 | | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae | | Descriptor: | Nicotinate phosphoribosyltransferase, SULFATE ION | | Authors: | Mulichak, A.M, Sauder, J.M, Keefe, L.J, Burley, S.K, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae
To be Published
|
|
4HYR
 
 | | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder
To be published
|
|
4HN8
 
 | | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp | | Descriptor: | D-glucarate dehydratase, GLYCEROL | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp
To be published
|
|
4I3V
 
 | |
4I3U
 
 | |
4I7B
 
 | | Siah1 bound to synthetic peptide (ACE)KLRPV(ABA)MVRPTVR | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ZINC ION | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I3W
 
 | |
4I7C
 
 | | Siah1 mutant bound to synthetic peptide (ACE)KLRPV(23P)MVRPWVR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I3T
 
 | |
6EXJ
 
 | | PDZ domain from rat Shank3 bound to the C terminus of somatostatin receptor subtype 2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3, SSTR2 | | Authors: | Ponna, S.K, Keller, C, Ruskamo, S, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
1WQG
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
1WQH
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structural studies of Mycobacterium Tuberculosis RRF and a comparative study of RRFS of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQF
 
 | | Crystal structure of Ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-28 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
