5QC9
 
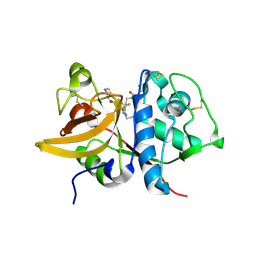 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-{[5-{1-[3-(dimethylamino)propyl]-5-[(piperidin-4-yl)acetyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}-2-(trifluoromethyl)phenyl]methyl}-3-methylbut-2-enamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCP
 
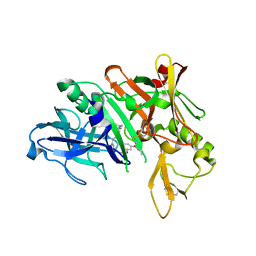 | | Crystal structure of BACE complex with BMC018 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6MPZ
 
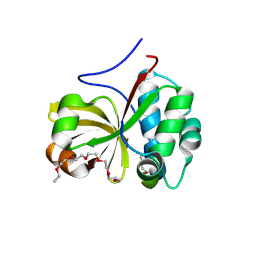 | | Crystal structure of a double glycine motif protease from AMS/PCAT transporter in complex with the leader peptide | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Double Glycine Motif Protease domain from AMS/PCAT Transporter, peptide aldehyde inhibitor 1 based on the ProcA2.8 leader peptide | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2018-10-09 | | Release date: | 2019-02-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease.
Elife, 8, 2019
|
|
5QD5
 
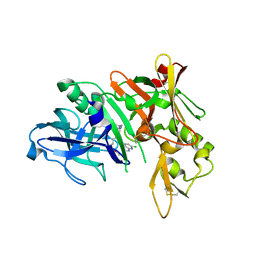 | | Crystal structure of BACE complex with BMC009 | | Descriptor: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
2IOJ
 
 | |
7QOS
 
 | |
2IPQ
 
 | | Crystal structure of C-terminal domain of Salmonella Enterica protein STY4665, PFAM DUF1528 | | Descriptor: | Hypothetical protein STY4665 | | Authors: | Ramagopal, U.A, Bonanno, J.B, Gilmore, J, Toro, R, Bain, K.T, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-terminal domain of Hypothetical protein STY4665
To be Published
|
|
2I3O
 
 | |
6SOE
 
 | | Mouse RBM20 RRM domain | | Descriptor: | RNA-binding protein 20 | | Authors: | Mackereth, C.D, Upadhyay, S.K. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis of UCUU RNA motif recognition by splicing factor RBM20.
Nucleic Acids Res., 48, 2020
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6MJG
 
 | |
6F94
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
2IJ9
 
 | | Crystal Structure of Uridylate Kinase from Archaeoglobus Fulgidus | | Descriptor: | SULFATE ION, Uridylate kinase | | Authors: | Patskovsky, Y, Chen, R, Keefe, L.J, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Uridylate Kinase from Archaeoglobus Fulgidus
To be Published
|
|
2IJZ
 
 | |
5OVV
 
 | | PDZ domain from rat Shank3 bound to the C terminus of ProSAPiP1 | | Descriptor: | Leucine zipper putative tumor suppressor 3, SH3 and multiple ankyrin repeat domains protein 3, THIOCYANATE ION | | Authors: | Ponna, S.K, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-08-29 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
5QC2
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-(3-hydroxypropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
2IF6
 
 | | Crystal structure of metalloprotein yiiX from Escherichia coli O157:H7, DUF1105 | | Descriptor: | Hypothetical protein yiiX, UNKNOWN ATOM OR ION | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical metalloprotein yiiX from Escherichia coli O157:H7
To be Published
|
|
5QCI
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, GLYCEROL, N-benzyl-1-{5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-2-methoxyphenyl}methanamine, ... | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | Descriptor: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD6
 
 | | Crystal structure of BACE complex with BMC004 | | Descriptor: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
2IMO
 
 | |
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5QCA
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{4-[(2-chloro-5-{1-[3-(4-cyclopropylpiperazin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}-N-[(4-chlorophenyl)methyl]methanamine, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
