3CZ8
 
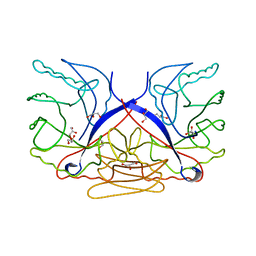 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3I6V
 
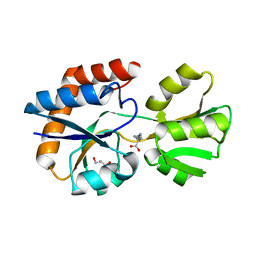 | | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine | | Descriptor: | GLYCEROL, LYSINE, SODIUM ION, ... | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine
To be Published
|
|
3HUR
 
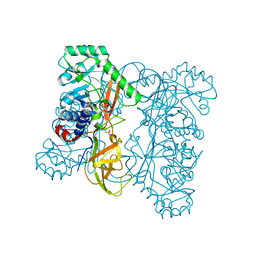 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, SULFATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine racemase from Oenococcus oeni
To be Published
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3L8K
 
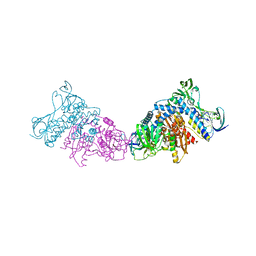 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
2R1F
 
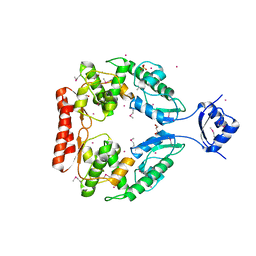 | | Crystal structure of predicted aminodeoxychorismate lyase from Escherichia coli | | Descriptor: | CADMIUM ION, GLYCEROL, Predicted aminodeoxychorismate lyase, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Rutter, M, Lau, C, Maletic, M, Smith, D, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Predicted Aminodeoxychorismate Lyase from Escherichia coli.
To be Published
|
|
3M9U
 
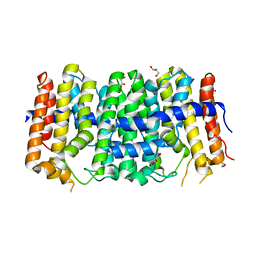 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Farnesyl-diphosphate synthase, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis Atcc 367
To be Published
|
|
2PTF
 
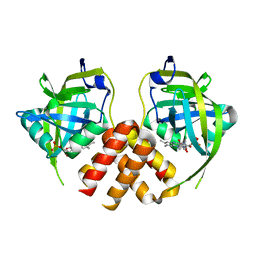 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
3BT5
 
 | | Crystal structure of DUF305 fragment from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Uncharacterized protein DUF305 | | Authors: | Ramagopal, U.A, Patskovsky, Y, Rutter, M, Toro, R, Bain, K, Meyer, A.J, Powell, A, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of DUF305 fragment from Deinococcus radiodurans.
To be Published
|
|
2P1J
 
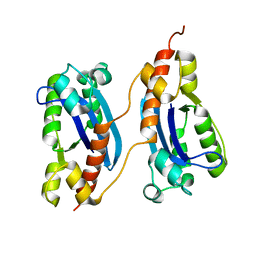 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
3CTD
 
 | | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris | | Descriptor: | Putative ATPase, AAA family | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris.
To be Published
|
|
3CLW
 
 | | Crystal structure of conserved exported protein from Bacteroides fragilis | | Descriptor: | Conserved exported protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of conserved exported protein from Bacteroides fragilis.
To be Published
|
|
3MZN
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | Descriptor: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
2P67
 
 | | Crystal structure of LAO/AO transport system kinase | | Descriptor: | CHLORIDE ION, LAO/AO transport system kinase, SODIUM ION | | Authors: | Ramagopal, U.A, Adams, J, Rodgers, L, Toro, R, Bain, K, Rutter, M, Schwinn, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LAO/AO transport system kinase
To be Published
|
|
2PCE
 
 | | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | PHOSPHATE ION, putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM
To be Published
|
|
2QSD
 
 | | Crystal structure of a protein Il1583 from Idiomarina loihiensis | | Descriptor: | GLYCEROL, Uncharacterized conserved protein | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Romero, R, Rutter, M, Koss, J, Mckenzie, C, Gheyi, T, Bain, K, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Protein Il1583 from Idiomarina loihiensis.
To be Published
|
|
3NIW
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Bonanno, J.B, Ramagopal, U, Toro, R, Rutter, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron
To be Published
|
|
2O16
 
 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
3N28
 
 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3N3D
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
2P2U
 
 | | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris | | Descriptor: | Host-nuclease inhibitor protein Gam, putative | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Zhang, F, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris
To be Published
|
|
3MY9
 
 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
3NFU
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | Descriptor: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
