3TQM
 
 | | Structure of an ribosomal subunit interface protein from Coxiella burnetii | | Descriptor: | Ribosome-associated factor Y, SULFATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQQ
 
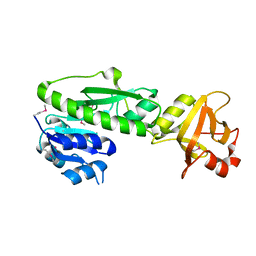 | | Structure of the methionyl-tRNA formyltransferase (fmt) from Coxiella burnetii | | Descriptor: | Methionyl-tRNA formyltransferase, POTASSIUM ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQO
 
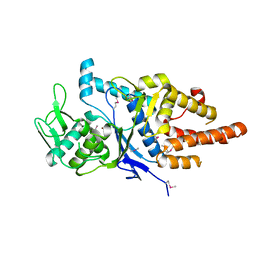 | | Structure of the cysteinyl-tRNA synthetase (cysS) from Coxiella burnetii. | | Descriptor: | Cysteinyl-tRNA synthetase, ZINC ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQV
 
 | | Structure of the nicotinate-nucleotide pyrophosphorylase from Francisella tularensis. | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase, PHOSPHATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Rapid countermeasure discovery against Francisella tularensis based on a metabolic network reconstruction.
Plos One, 8, 2013
|
|
3TQS
 
 | | Structure of the dimethyladenosine transferase (ksgA) from Coxiella burnetii | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3OLF
 
 | |
3OOF
 
 | |
3OOK
 
 | |
3OKI
 
 | |
3OMM
 
 | |
3OKH
 
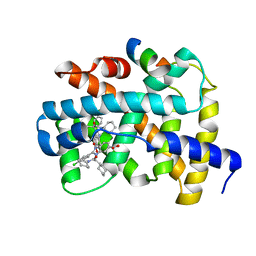 | |
3OMK
 
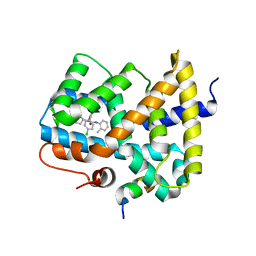 | |
1Z70
 
 | | 1.15A resolution structure of the formylglycine generating enzyme FGE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-alpha-formyglycine-generating enzyme, ... | | Authors: | Rudolph, M.G. | | Deposit date: | 2005-03-23 | | Release date: | 2005-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | De novo calcium/sulfur SAD phasing of the human formylglycine-generating enzyme using in-house data.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3NBF
 
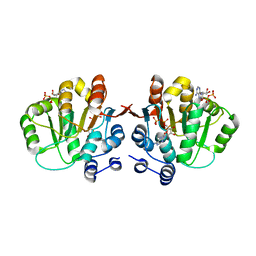 | | Q28E mutant of hera helicase N-terminal domain bound to 8-oxo-ADP | | Descriptor: | Heat resistant RNA dependent ATPase, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
4DDX
 
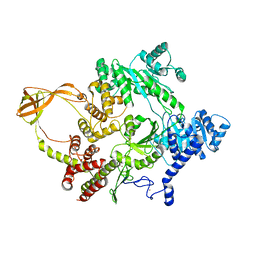 | |
4DDU
 
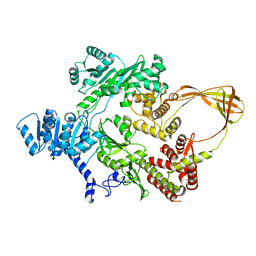 | | Thermotoga maritima reverse gyrase, C2 FORM 1 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDT
 
 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDW
 
 | |
4DDV
 
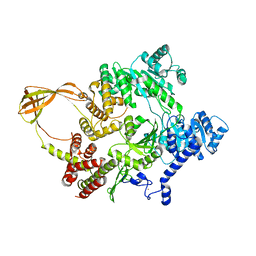 | |
3OIY
 
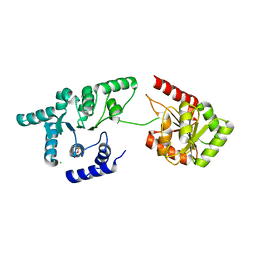 | | Helicase domain of reverse gyrase from Thermotoga maritima | | Descriptor: | CHLORIDE ION, PYROPHOSPHATE 2-, reverse gyrase helicase domain | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The latch modulates nucleotide and DNA binding to the helicase-like domain of Thermotoga maritima reverse gyrase and is required for positive DNA supercoiling.
Nucleic Acids Res., 39, 2011
|
|
3MWL
 
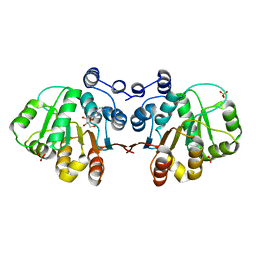 | | Q28E mutant of HERA N-terminal RecA-like domain in complex with 8-OXOADENOSINE | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-7H-purin-8-one, Heat resistant RNA dependent ATPase, SULFATE ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3MWJ
 
 | |
3MWK
 
 | | Q28E mutant of HERA N-terminal RecA-like domain, complex with 8-oxo-AMP | | Descriptor: | Heat resistant RNA dependent ATPase, SULFATE ION, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3NEJ
 
 | |
3P4Y
 
 | |
