7TK7
 
 | |
7TK4
 
 | |
7TKC
 
 | |
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG6
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG5
 
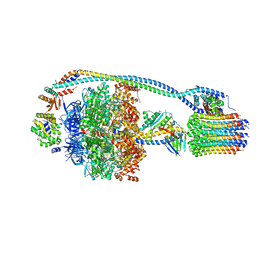 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGC
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG9
 
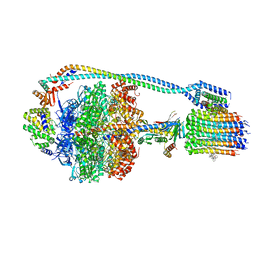 | | Cryo-EM structure of bedaquiline-saturated mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG7
 
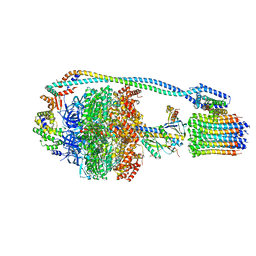 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
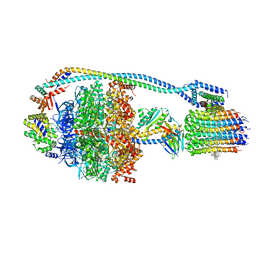 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGB
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7TAP
 
 | | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | | Descriptor: | Archazolid A, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7TAO
 
 | | Cryo-EM structure of bafilomycin A1 bound to yeast VO V-ATPase | | Descriptor: | (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7TMQ
 
 | | V1 complex lacking subunit C from Saccharomyces cerevisiae, State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, MAGNESIUM ION, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMP
 
 | | V1 complex lacking subunit C from Saccharomyces cerevisiae, State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, MAGNESIUM ION, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMM
 
 | | Complete V1 Complex from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMO
 
 | | V1 complex lacking subunit C from Saccharomyces cerevisiae, State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, H(+)-transporting two-sector ATPase, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMT
 
 | | V-ATPase from Saccharomyces cerevisiae, State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMR
 
 | | V-ATPase from Saccharomyces cerevisiae, State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMS
 
 | | V-ATPase from Saccharomyces cerevisiae, State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TJW
 
 | |
7TJX
 
 | |
7TJT
 
 | |
7TJS
 
 | |
7TJU
 
 | |
