1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
4WY9
 
 | |
1CWU
 
 | | BRASSICA NAPUS ENOYL ACP REDUCTASE A138G MUTANT COMPLEXED WITH NAD+ AND THIENODIAZABORINE | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ENOYL ACP REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Roujeinikova, A, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor binding studies on enoyl reductase reveal conformational changes related to substrate recognition.
J.Biol.Chem., 274, 1999
|
|
4RI1
 
 | |
2Q7Q
 
 | |
1LWH
 
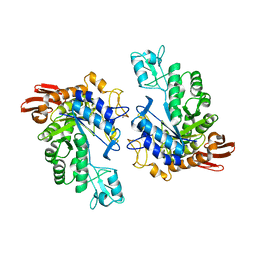 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1L0I
 
 | | Crystal structure of butyryl-ACP I62M mutant | | Descriptor: | Acyl carrier protein, CACODYLATE ION, SODIUM ION, ... | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray Crystallographic Studies on Butyryl-ACP Reveal Flexibility of the Structure around a Putative Acyl Chain Binding Site
Structure, 10, 2002
|
|
1L0H
 
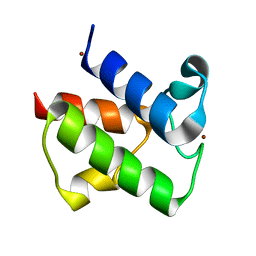 | | CRYSTAL STRUCTURE OF BUTYRYL-ACP FROM E.COLI | | Descriptor: | ACYL CARRIER PROTEIN, ZINC ION | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site
Structure, 10, 2002
|
|
1LWJ
 
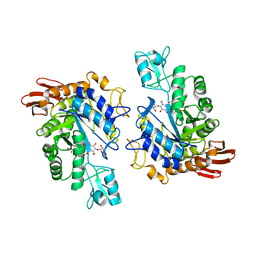 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE/ACARBOSE COMPLEX | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, MODIFIED ACARBOSE PENTASACCHARIDE | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1GJW
 
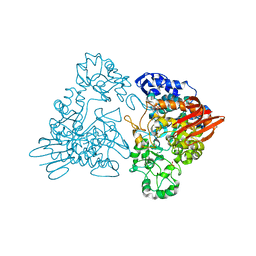 | | Thermotoga maritima maltosyltransferase complex with maltose | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
1GJU
 
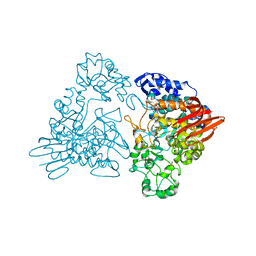 | | Maltosyltransferase from Thermotoga maritima | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-02 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
3IMP
 
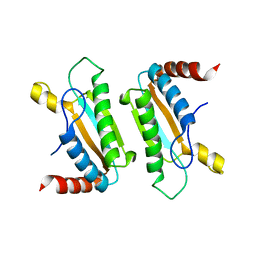 | |
4XMR
 
 | | Crystal structure of the sensory domain of the Campylobacter jejuni chemoreceptor Tlp3 (CcmL) with isoleucine bound. | | Descriptor: | ISOLEUCINE, Putative methyl-accepting chemotaxis signal transduction protein, SULFATE ION | | Authors: | Roujeinikova, A, Liu, Y.C, Machuca, M.A. | | Deposit date: | 2015-01-15 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XMQ
 
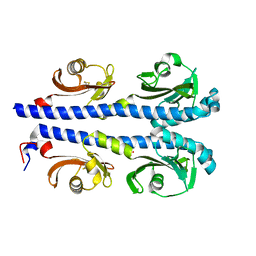 | |
4YHA
 
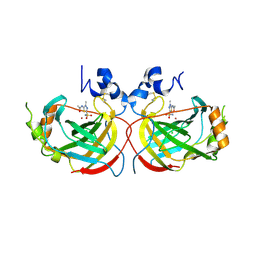 | |
3DPQ
 
 | |
4YGF
 
 | | Crystal structure of the complex of Helicobacter pylori alpha-Carbonic Anhydrase with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Roujeinikova, A, Modak, J.K. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase by Sulfonamides.
Plos One, 10, 2015
|
|
3DPO
 
 | |
3CYP
 
 | |
3DPP
 
 | |
3R6M
 
 | | Crystal structure of Vibrio parahaemolyticus YeaZ | | Descriptor: | YeaZ, resuscitation promoting factor | | Authors: | Roujeinikova, A, Aydin, I. | | Deposit date: | 2011-03-21 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Analysis of the Essential Resuscitation Promoting Factor YeaZ Suggests a Mechanism of Nucleotide Regulation through Dimer Reorganization.
Plos One, 6, 2011
|
|
2IUR
 
 | |
2IUV
 
 | |
2OK6
 
 | |
2OIZ
 
 | |
