6XR6
 
 | |
6XR7
 
 | |
6XRG
 
 | |
7MZY
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
2KCV
 
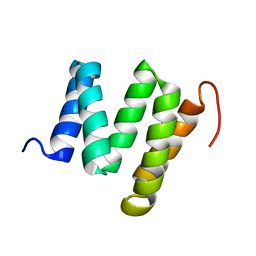 | | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c
To be Published
|
|
2KCP
 
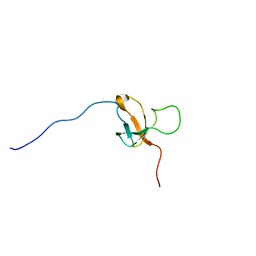 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E; FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET Tr71d | | Descriptor: | 30S ribosomal protein S8e | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-24 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET
To be Published
|
|
2KEM
 
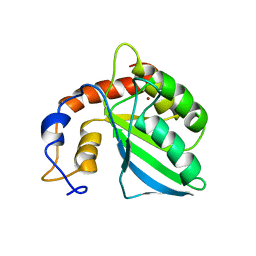 | | Extended structure of citidine deaminase domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Harjes, E, Gross, P.J, Chen, K, Lu, Y, Shindo, K, Nowarski, R, Gross, J.D, Kotler, M, Harris, R.S, Matsuo, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model
J.Mol.Biol., 389, 2009
|
|
4UMC
 
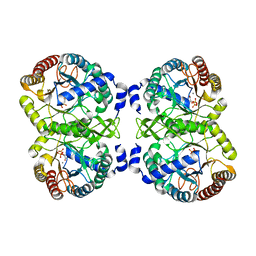 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMB
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
3IPF
 
 | | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR8c. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Andre, I, Rossi, P, Kennedy, M, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
6AMW
 
 | |
6AMV
 
 | |
3LMO
 
 | | Crystal Structure of specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Forouhar, F, Rossi, P, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
2MS4
 
 | | Cyclophilin a complexed with a fragment of crk-ii | | Descriptor: | Peptide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Jankowski, W, Saleh, T, Rossi, P, Kalodimos, C. | | Deposit date: | 2014-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway.
Nat.Chem.Biol., 12, 2016
|
|
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
6PPT
 
 | |
5JTM
 
 | | The structure of chaperone SecB in complex with unstructured PhoA binding site a | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTQ
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site d | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
6PQ2
 
 | |
6PQM
 
 | |
5JTP
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site e | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTO
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site d | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTR
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site e | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
