3IHM
 
 | |
1MTY
 
 | | METHANE MONOOXYGENASE HYDROXYLASE FROM METHYLOCOCCUS CAPSULATUS (BATH) | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Rosenzweig, A.C, Nordlund, P, Lippard, S.J, Frederick, C.A. | | Deposit date: | 1996-07-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions.
Proteins, 29, 1997
|
|
4KNT
 
 | | Copper nitrite reductase from Nitrosomonas europaea pH 8.5 | | Descriptor: | COPPER (II) ION, GLYCEROL, Multicopper oxidase type 1 | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
4KNU
 
 | | Copper nitrite reductase from Nitrosomonas europaea at pH 6.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
4KNS
 
 | | Reduced crystal structure of the Nitrosomonas europaea copper nitrite reductase at pH 6.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
1MMO
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL NON-HAEM IRON HYDROXYLASE THAT CATALYSES THE BIOLOGICAL OXIDATION OF METHANE | | Descriptor: | ACETIC ACID, FE (III) ION, METHANE MONOOXYGENASE HYDROLASE (ALPHA CHAIN), ... | | Authors: | Rosenzweig, A.C, Frederick, C.A, Lippard, S.J, Nordlund, P. | | Deposit date: | 1994-02-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane.
Nature, 366, 1993
|
|
1CC8
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, MERCURY (II) ION, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CC7
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
4N83
 
 | |
2PS0
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ZINC ION | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
6P1F
 
 | | apo PmoF2 PCuAC domain | | Descriptor: | Copper chaperone PCu(A)C | | Authors: | Fisher, O.S, Sendzik, M.R, Rosenzweig, A.C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | PCuAC domains from methane-oxidizing bacteria use a histidine brace to bind copper.
J.Biol.Chem., 294, 2019
|
|
8OYI
 
 | | particulate methane monooxygenase with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
5C91
 
 | | NEDD4 HECT with covalently bound indole-based inhibitor | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | | Authors: | Span, I, Smith, A.T, Kathman, S, Statsyuk, A.V, Rosenzweig, A.C. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism.
J. Am. Chem. Soc., 137, 2015
|
|
1YEW
 
 | |
1AIO
 
 | | CRYSTAL STRUCTURE OF A DOUBLE-STRANDED DNA CONTAINING THE MAJOR ADDUCT OF THE ANTICANCER DRUG CISPLATIN | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(BRU)P*CP*TP*[PT(NH3)2(GP*GP)]*TP*CP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3') | | Authors: | Takahara, P.M, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 1997-04-23 | | Release date: | 1997-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin.
Nature, 377, 1995
|
|
7N0J
 
 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
2ROP
 
 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
4ZAJ
 
 | |
4YDX
 
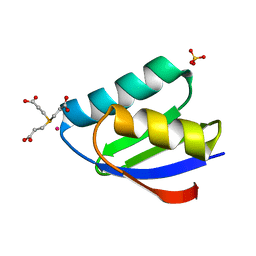 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7L6G
 
 | | MbnP from Methylosinus trichosporium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Manesis, A.C, Rosenzweig, A.C. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Copper binding by a unique family of metalloproteins is dependent on kynurenine formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SR4
 
 | | particulate methane monooxygeanse treated with potassium cyanide and copper reloaded | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (II) ION, ... | | Authors: | Tucci, F.J, Jodts, R.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analogue binding identifies the copper active site of particulate methane monooyxgenase
Nat Catal, 2023
|
|
8SR1
 
 | | particulate methane monooxygenase crosslinked with 4,4,4-trifluorobutanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 4,4,4-trifluorobutan-1-ol, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SQW
 
 | | particulate methane monooxygenase crosslinked with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR5
 
 | |
