1RG5
 
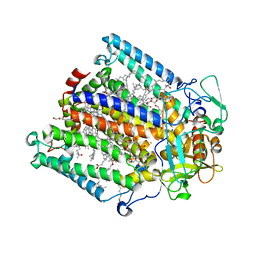 | | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
1GJH
 
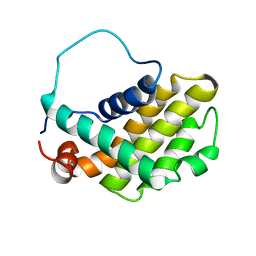 | | HUMAN BCL-2, ISOFORM 2 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3SS5
 
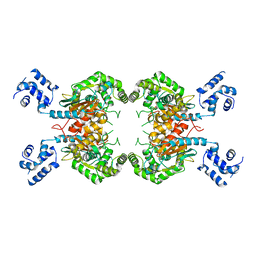 | |
1G5M
 
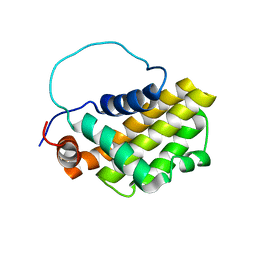 | | HUMAN BCL-2, ISOFORM 1 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3SS4
 
 | |
1GMX
 
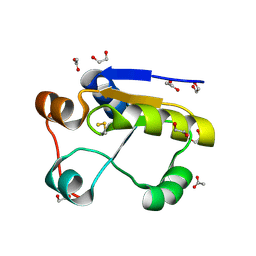 | | Escherichia coli GlpE sulfurtransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
5EKF
 
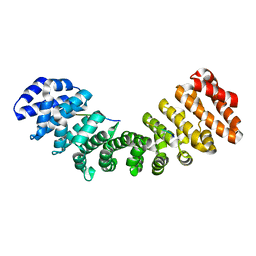 | |
3SS3
 
 | |
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1H05
 
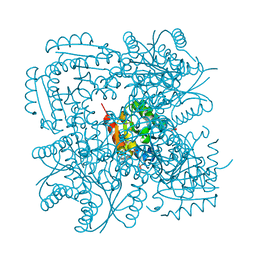 | |
1HM6
 
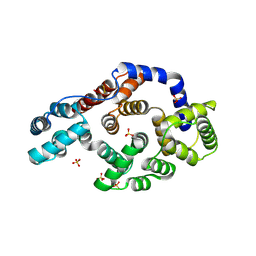 | |
5EKG
 
 | |
3T6D
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-08 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
1H0S
 
 | | 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with 3-hydroxyimino-quinic acid | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, 3-HYDROXYIMINO QUINIC ACID, GLYCEROL, ... | | Authors: | Roszak, A.W, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Specificity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1H0R
 
 | | Type II Dehydroquinase from Mycobacterium tuberculosis complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Roszak, A.W, Robinson, D.A, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selectivity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
3T6E
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-94 | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
2VG5
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
5U5R
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus musculus Complexed with a PMS2 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Importin subunit alpha-1, Mismatch repair endonuclease PMS2 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
1URH
 
 | | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli | | Descriptor: | 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Spallarossa, A, Forlani, F, Carpen, A, Armirotti, A, Pagani, S, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-10-30 | | Release date: | 2003-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The "Rhodanese" Fold and Catalytic Mechanism of 3-Mercaptopyruvate Sulfurtransferases: Crystal Structure of Ssea from Escherichia Coli
J.Mol.Biol., 335, 2004
|
|
1OOZ
 
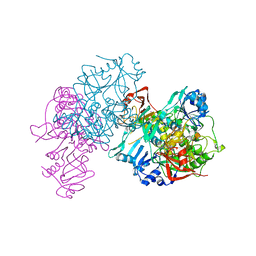 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3UVU
 
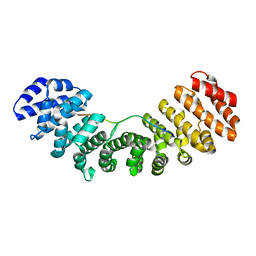 | |
4FZL
 
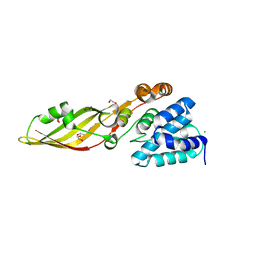 | | High resolution structure of truncated bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION, ... | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
6PPW
 
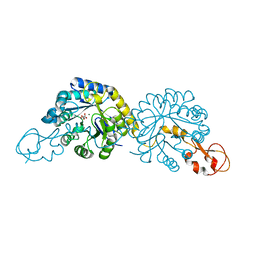 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with magnesium and malate | | Descriptor: | D-MALATE, MAGNESIUM ION, N-acetylneuraminate synthase | | Authors: | Rosanally, A.Z, Junop, M.S, Berti, P.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
