3FAH
 
 | | Glycerol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Aldehyde oxidoreductase, CHLORIDE ION, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
3FC4
 
 | | Ethylene glycol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 1,2-ETHANEDIOL, Aldehyde oxidoreductase, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
2WYS
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-20 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
3KAP
 
 | | Flavodoxin from Desulfovibrio desulfuricans ATCC 27774 (oxidized form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
1RZ5
 
 | | Di-haem Cytochrome c Peroxidase, Form OUT | | Descriptor: | CALCIUM ION, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A.S, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
1RZ6
 
 | | Di-haem Cytochrome c Peroxidase, Form IN | | Descriptor: | CITRIC ACID, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
3ERX
 
 | | High-resolution structure of Paracoccus pantotrophus pseudoazurin | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Najmudin, S, Pauleta, S.R, Moura, I, Romao, M.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3KAQ
 
 | | Flavodoxin from D. desulfuricans (semireduced form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
2P1E
 
 | | Crystal structure of the Leishmania infantum glyoxalase II with D-Lactate at the active site | | Descriptor: | Glyoxalase II, LACTIC ACID, SPERMIDINE, ... | | Authors: | Trincao, J, Barata, L, Najmudin, S, Bonifacio, C, Romao, M.J. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Structural Properties of Leishmania infantum Glyoxalase II: Trypanothione Specificity and Phylogeny.
Biochemistry, 47, 2008
|
|
6FU3
 
 | | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | CALCIUM ION, HEME C, Protein CcpR | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
2P18
 
 | | Crystal structure of the Leishmania infantum glyoxalase II | | Descriptor: | ACETIC ACID, Glyoxalase II, SPERMIDINE, ... | | Authors: | Trincao, J, Barata, L, Najmudin, S, Bonifacio, C, Romao, M.J. | | Deposit date: | 2007-03-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalysis and Structural Properties of Leishmania infantum Glyoxalase II: Trypanothione Specificity and Phylogeny.
Biochemistry, 47, 2008
|
|
3L0P
 
 | | Crystal structures of Iron containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | Adenylate kinase, FE (III) ION, GLYCEROL | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
8RC9
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 2 h | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RC8
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 0 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCB
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air (in a not degassed drop) for 34 min in the presence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCC
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - aerobic soaked with 48 bar CO2 for 1 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBON DIOXIDE, ... | | Authors: | Vilela-Alves, G, Carpentier, P, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCA
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air for 1 h in the absence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
2WFB
 
 | | High resolution structure of the apo form of the orange protein (ORP) from Desulfovibrio gigas | | Descriptor: | ACETATE ION, PHOSPHATE ION, PUTATIVE UNCHARACTERIZED PROTEIN ORP | | Authors: | Najmudin, S, Bonifacio, C, Duarte, A.G, Pereira, A.S, Moura, I, Moura, J.M, Romao, M.J. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Apo Form of the Orange Protein (Apo-Orp) from Desulfovibrio Gigas
To be Published
|
|
8RCG
 
 | | W-formate dehydrogenase M405S from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2C4X
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
8QTY
 
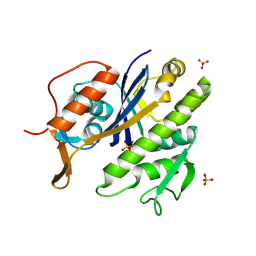 | |
1V0A
 
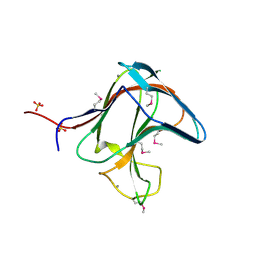 | | Family 11 Carbohydrate-Binding Module of cellulosomal cellulase Lic26A-Cel5E of Clostridium thermocellum | | Descriptor: | CALCIUM ION, ENDOGLUCANASE H, SULFATE ION | | Authors: | Carvalho, A.L, Romao, M.J, Goyal, A, Prates, J.A.M, Pires, V.M.R, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accomodates Beta-1,4- and Beta-1,3-1,4-Mixed Linked Glucans at a Single Binding Site
J.Biol.Chem., 279, 2004
|
|
2LK5
 
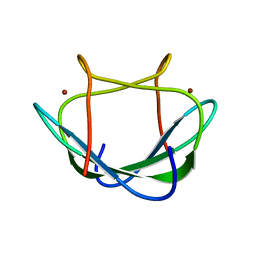 | | Solution structure of the Zn(II) form of Desulforedoxin | | Descriptor: | Desulforedoxin, ZINC ION | | Authors: | Goodfellow, B.J, Tavares, P, Romao, M.J, Czaja, C, Rusnak, F, Legall, J, Moura, I, Moura, J.J.G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of desulforedoxin, a simple iron-sulfur protein - An NMR study of the zinc derivative
J.BIOL.INORG.CHEM., 1, 1996
|
|
2NAP
 
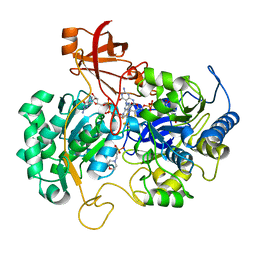 | | DISSIMILATORY NITRATE REDUCTASE (NAP) FROM DESULFOVIBRIO DESULFURICANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Dias, J.M, Than, M, Humm, A, Huber, R, Bourenkov, G, Bartunik, H, Bursakov, S, Calvete, J, Caldeira, J, Carneiro, C, Moura, J, Moura, I, Romao, M.J. | | Deposit date: | 1998-09-18 | | Release date: | 1999-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods.
Structure Fold.Des., 7, 1999
|
|
