6M0R
 
 | | 2.7A Yeast Vo state3 | | Descriptor: | (6~{E},10~{E},14~{E},18~{E},22~{E},26~{E},30~{R})-2,6,10,14,18,22,26,30-octamethyldotriaconta-2,6,10,14,18,22,26-heptaene, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, PYROPHOSPHATE, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, Singharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
6M0S
 
 | | 3.6A Yeast Vo state3 prime | | Descriptor: | Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, SIngharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
8I9Q
 
 | | The focused refinement of CCT4-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | Phosducin-like protein 3, T-complex protein 1 subunit delta | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8IB8
 
 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I9U
 
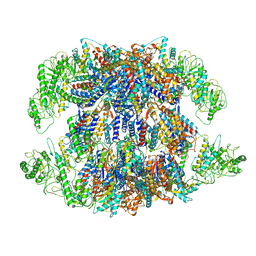 | | Human TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I1U
 
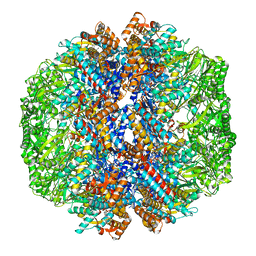 | | Human TRiC-PhLP2A complex in the closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I6J
 
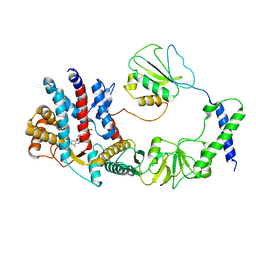 | | The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit gamma | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
5W0S
 
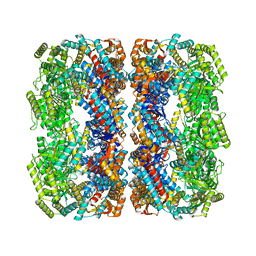 | | GroEL using cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Roh, S.H, Chiu, W. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Subunit conformational variation within individual GroEL oligomers resolved by Cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7E5Z
 
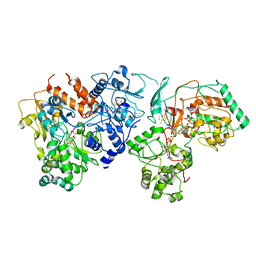 | | Dehydrogenase holoenzyme | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Roh, S.H, Park, J.S, Heo, Y.Y. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dehydrogenase holoenzyme
To Be Published
|
|
1LA4
 
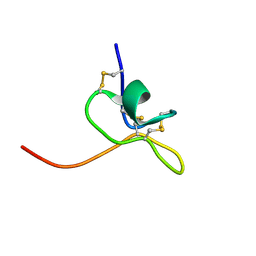 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
6NRA
 
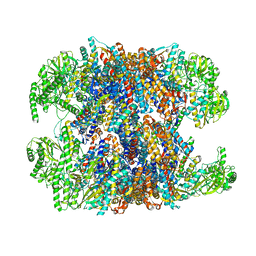 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRC
 
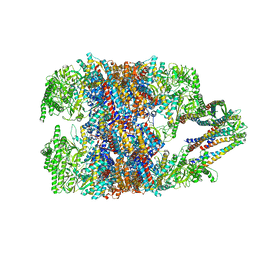 | | hTRiC-hPFD Class3 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NR9
 
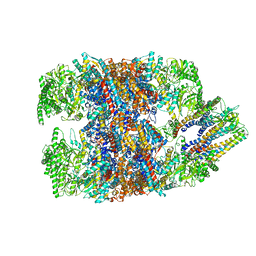 | | hTRiC-hPFD Class5 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRD
 
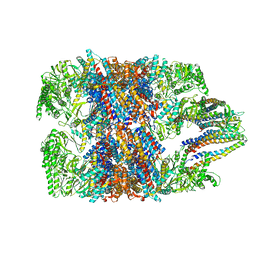 | | hTRiC-hPFD Class4 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRB
 
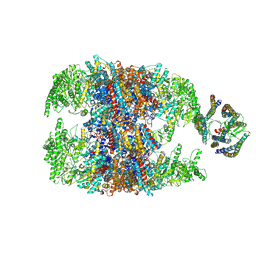 | | hTRiC-hPFD Class2 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NR8
 
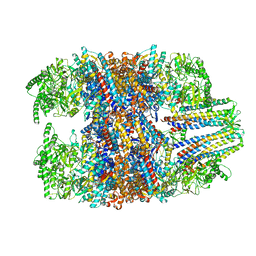 | | hTRiC-hPFD Class6 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6CNK
 
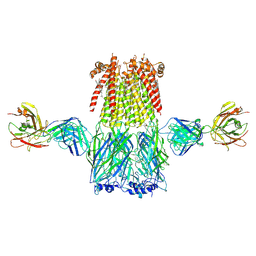 | | Structure of the 3alpha2beta stiochiometry of the human Alpha4Beta2 nicotinic receptor | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Walsh Jr, R.M, Roh, S.H, Gharpure, A, Morales-Perez, C.L, Hibbs, R.E. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural principles of distinct assemblies of the human alpha 4 beta 2 nicotinic receptor.
Nature, 557, 2018
|
|
6CNJ
 
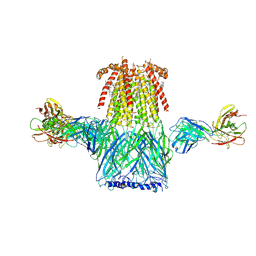 | | Structure of the 2alpha3beta stiochiometry of the human Alpha4Beta2 nicotinic receptor | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Walsh Jr, R.M, Roh, S.H, Gharpure, A, Morales-Perez, C.L, Teng, J, Hibbs, R.E. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural principles of distinct assemblies of the human alpha 4 beta 2 nicotinic receptor.
Nature, 557, 2018
|
|
7VWX
 
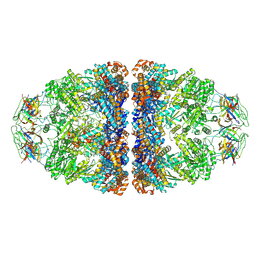 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | Authors: | Kim, H, Roh, S.H. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
7XQW
 
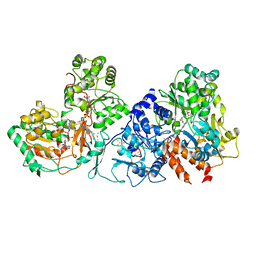 | | Formate dehydrogenase (FDH) from Methylobacterium extorquens AM1 (MeFDH1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Park, J, Heo, Y.Y, Roh, S.H, Lee, H.H. | | Deposit date: | 2022-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Enzymatic conversion of CO2 in real flue gas to molar-scale formate
To Be Published
|
|
7FDA
 
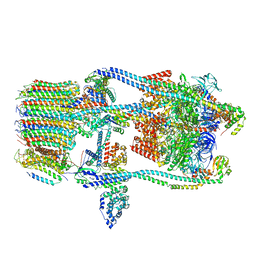 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
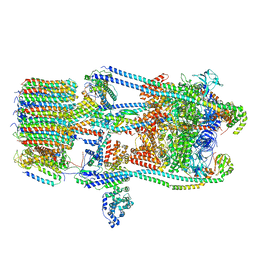 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDB
 
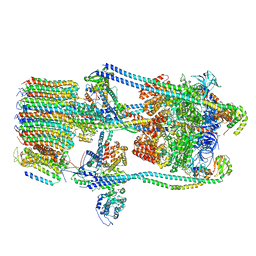 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
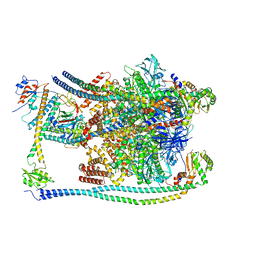 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
