5AFU
 
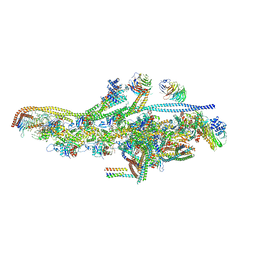 | | Cryo-EM structure of dynein tail-dynactin-BICD2N complex | | Descriptor: | ACTIN, CYTOPLASMIC 1, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Urnavicius, L, Zhang, K, Diamant, A.G, Motz, C, Schlager, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-01-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
2VRD
 
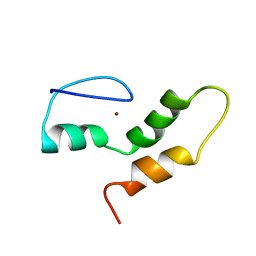 | | THE STRUCTURE OF THE ZINC FINGER FROM THE HUMAN SPLICEOSOMAL PROTEIN U1C | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN C, ZINC ION | | Authors: | Muto, Y, Pomeranz-Krummel, D, Oubridge, C, Hernandez, H, Robinson, C, Neuhaus, D, Nagai, K. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure and Biochemical Properties of the Human Spliceosomal Protein U1C
J.Mol.Biol., 341, 2004
|
|
2BYU
 
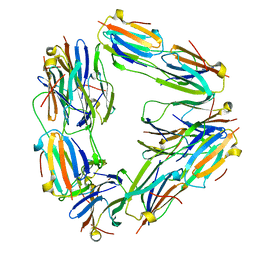 | | Negative stain EM reconstruction of M.tuberculosis Acr1(Hsp 16.3) fitted with wheat sHSP dimer | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Kennaway, C.K, Benesch, J.L.P, Gohlke, U, Wang, L, Robinson, C.V, Orlova, E.V, Saibil, H.R, Keep, N.H. | | Deposit date: | 2005-08-05 | | Release date: | 2005-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dodecameric Structure of the Small Heat Shock Protein Acr1 from Mycobacterium Tuberculosis.
J.Biol.Chem., 280, 2005
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
3D36
 
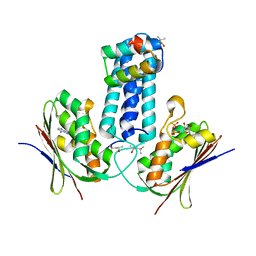 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
5TJ5
 
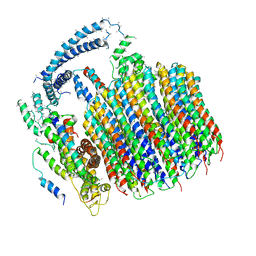 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
4NH2
 
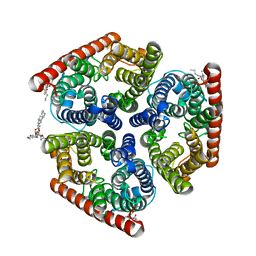 | | Crystal structure of AmtB from E. coli bound to phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Ammonia channel | | Authors: | Laganowsky, A, Reading, E, Allison, T.M, Robinson, C.V. | | Deposit date: | 2013-11-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membrane proteins bind lipids selectively to modulate their structure and function.
Nature, 510, 2014
|
|
2DOQ
 
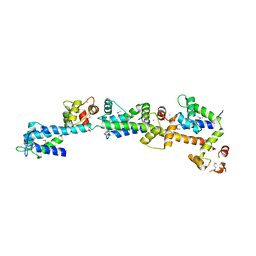 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
5FTT
 
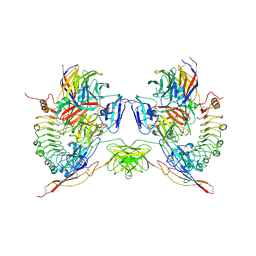 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
4CYJ
 
 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
2GV5
 
 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | Cell division control protein 31, Sfi1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
4CYI
 
 | | Chaetomium thermophilum Pan3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
1VAZ
 
 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
2P1P
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1O
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, NAPHTHALEN-1-YL-ACETIC ACID, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
1V92
 
 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
2P1M
 
 | | TIR1-ASK1 complex structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, TRANSPORT INHIBITOR RESPONSE 1 protein | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-05 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
4M5T
 
 | | Disulfide trapped human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Cascio, D, Hochberg, G, Sawaya, M.R, Benesch, J.L.P, Robinson, C.V, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2P1Q
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase.
Nature, 446, 2007
|
|
2JAP
 
 | | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the beta-Lactamase Inhibitor Clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, CLAVALDEHYDE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
1OP9
 
 | | Complex of human lysozyme with camelid VHH HL6 antibody fragment | | Descriptor: | HL6 camel VHH fragment, Lysozyme C | | Authors: | Dumoulin, M, Last, A.M, Desmyter, A, Decanniere, K, Canet, D, Larsson, G, Spencer, A, Archer, D.B, Sasse, J, Muyldermans, S, Wyns, L, Redfield, C, Matagne, A, Robinson, C.V, Dobson, C.M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme
Nature, 424, 2003
|
|
5G2E
 
 | | Structure of the Nap1 H2A H2B complex | | Descriptor: | HISTONE H2A TYPE 1, HISTONE H2B 1.1, NUCLEOSOME ASSEMBLY PROTEIN | | Authors: | AguilarGurrieri, C, Larabi, A, Vinayachandran, V, Patel, N.A, Yen, K, Reja, R, Ebong, I.O, Schoehn, G, Robinson, C.V, Pugh, B.F, Panne, D. | | Deposit date: | 2016-04-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structural Evidence for Nap1-Dependent H2A-H2B Deposition and Nucleosome Assembly.
Embo J., 35, 2016
|
|
2JAH
 
 | | Biochemical and structural analysis of the Clavulanic acid dehydeogenase (CAD) from Streptomyces clavuligerus | | Descriptor: | CLAVULANIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
