5M5C
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | 分子名称: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | 登録日 | 2016-10-21 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NJL
 
 | | Cwp2 from Clostridium difficile | | 分子名称: | Cell surface protein (Putative S-layer protein), SULFATE ION | | 著者 | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2017-03-29 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro.
FEBS J., 284, 2017
|
|
4D59
 
 | | Clostridial Cysteine protease Cwp84 C116A after propeptide cleavage | | 分子名称: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | 著者 | Bradshaw, W.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2014-11-03 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Cwp84, a Clostridium Difficile Cysteine Protease, Exhibits Conformational Flexibility in the Absence of its Propeptide
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4D5A
 
 | | Clostridial Cysteine protease Cwp84 C116A after propeptide cleavage | | 分子名称: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), GLYCEROL, ... | | 著者 | Bradshaw, W.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2014-11-03 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cwp84, a Clostridium Difficile Cysteine Protease, Exhibits Conformational Flexibility in the Absence of its Propeptide
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4CI7
 
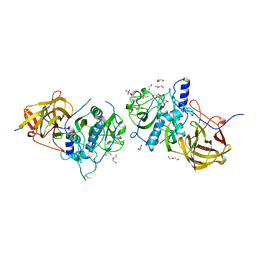 | | The crystal structure of the cysteine protease and lectin-like domains of Cwp84, a surface layer associated protein of Clostridium difficile | | 分子名称: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), GLYCEROL, ... | | 著者 | Bradshaw, W.J, Kirby, J.M, Thiyagarajan, N, Chambers, C.J, Davies, A.H, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Structure of the Cysteine Protease and Lectin-Like Domains of Cwp84, a Surface Layer-Associated Protein from Clostridium Difficile
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1KLA
 
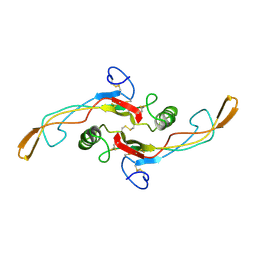 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | 分子名称: | TRANSFORMING GROWTH FACTOR-BETA 1 | | 著者 | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | 登録日 | 1996-01-16 | | 公開日 | 1996-08-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLC
 
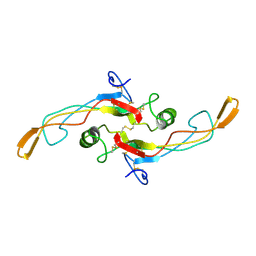 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | TRANSFORMING GROWTH FACTOR-BETA 1 | | 著者 | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | 登録日 | 1996-01-16 | | 公開日 | 1996-08-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLD
 
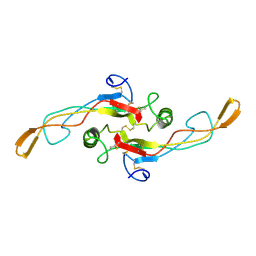 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | 分子名称: | TRANSFORMING GROWTH FACTOR-BETA 1 | | 著者 | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | 登録日 | 1996-01-16 | | 公開日 | 1996-08-17 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
2WN5
 
 | |
2WN7
 
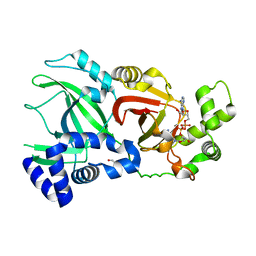 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | 分子名称: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2009-07-07 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2WN8
 
 | |
2WN4
 
 | |
2WN6
 
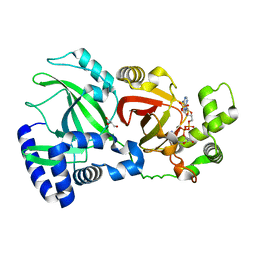 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | 分子名称: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | 登録日 | 2009-07-07 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2W05
 
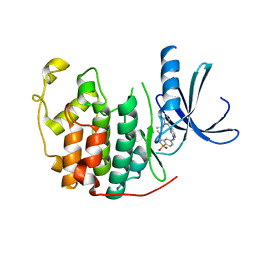 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5b | | 分子名称: | CELL DIVISION PROTEIN KINASE 2, N-(2-METHOXYETHYL)-4-({4-[2-METHYL-1-(1-METHYLETHYL)-1H-IMIDAZOL-5-YL]PYRIMIDIN-2-YL}AMINO)BENZENESULFONAMIDE | | 著者 | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Breed, J, Byth, K.F, Culshaw, J.D, Finlay, M.R, Fisher, E, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Pauptit, R.A, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | 登録日 | 2008-08-08 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VV9
 
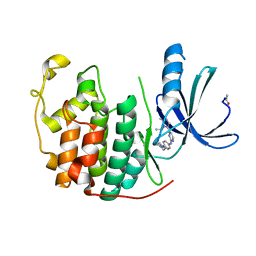 | | CDK2 in complex with an imidazole piperazine | | 分子名称: | 2-{4-[4-({4-[2-methyl-1-(1-methylethyl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)phenyl]piperazin-1-yl}-2-oxoethanol, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Acton, D.G, Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Finlay, M.R, Fisher, E, Gerhardt, S, Graham, M.A, Green, C.P, Heaton, D.W, Loddick, S.A, Morgentin, R, Read, J, Roberts, A, Stanway, J, Tucker, J.A, Weir, H.M. | | 登録日 | 2008-06-04 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Imidazole Piperazines: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors with a Novel Binding Mode.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2W06
 
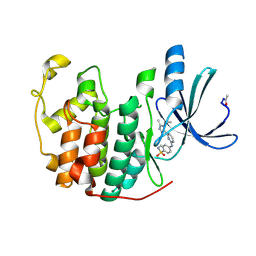 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5c | | 分子名称: | 4-{[4-(1-CYCLOPROPYL-2-METHYL-1H-IMIDAZOL-5-YL)PYRIMIDIN-2-YL]AMINO}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Byth, K.F, Culshaw, J.D, Finlay, M.R.V, Fisher, E, Mcmiken, H.H.J, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | 登録日 | 2008-08-08 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1HE7
 
 | | Human Nerve growth factor receptor TrkA | | 分子名称: | GLYCEROL, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | 著者 | Banfield, M, Robertson, A, Allen, S, Dando, J, Tyler, S, Bennett, G, Brain, S, Mason, G, Holden, P, Clarke, A, Naylor, R, Wilcock, G, Brady, R, Dawbarn, D. | | 登録日 | 2000-11-20 | | 公開日 | 2001-04-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification and Structure of the Nerve Growth Factor Binding Site on Trka.
Biochem.Biophys.Res.Commun., 282, 2001
|
|
6EMN
 
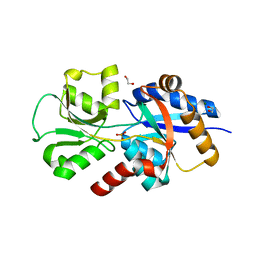 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | 著者 | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | 登録日 | 2017-10-03 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | 分子名称: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | 著者 | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | 登録日 | 2021-04-13 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
6YDL
 
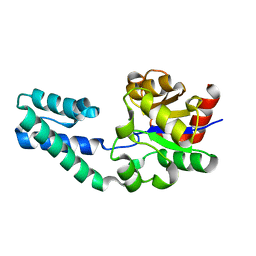 | | Substrate-free beta-phosphoglucomutase from Lactococcus lactis | | 分子名称: | Beta-phosphoglucomutase, MAGNESIUM ION | | 著者 | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | 登録日 | 2020-03-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6YDK
 
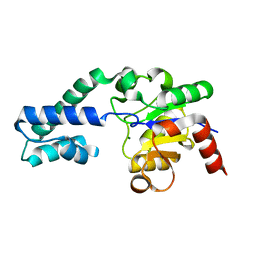 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | 分子名称: | Beta-phosphoglucomutase, MAGNESIUM ION | | 著者 | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | 登録日 | 2020-03-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6NJU
 
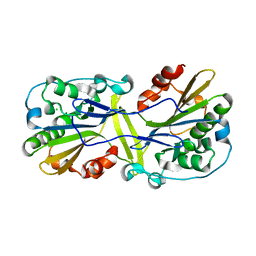 | | Mouse endonuclease G mutant H97A bound to A-DNA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DNA (5'-D(CCGGCGCCGG)-3'), ... | | 著者 | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | 登録日 | 2019-01-04 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
6YDJ
 
 | | P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with glucose 6-phosphate and trifluoromagnesate | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | 著者 | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | 登録日 | 2020-03-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6YDM
 
 | | beta-phosphoglucomutase from Lactococcus lactis with citrate, tris and acetate bound | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | 登録日 | 2020-03-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6NJT
 
 | | Mouse endonuclease G mutant - H97A | | 分子名称: | CHLORIDE ION, Endonuclease G, mitochondrial, ... | | 著者 | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | 登録日 | 2019-01-04 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
