3ZHN
 
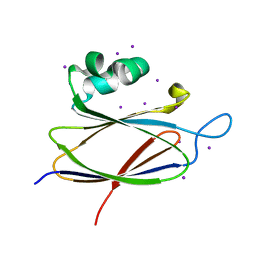 | | Crystal structure of the T6SS lipoprotein TssJ1 from Pseudomonas aeruginosa | | Descriptor: | IODIDE ION, PA_0080 | | Authors: | Robb, C.S, Assmus, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-12-22 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the T6Ss Lipoprotein Tssj1 from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ACL
 
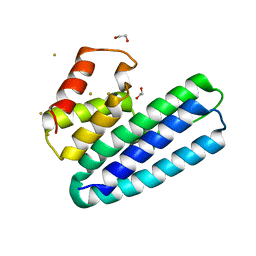 | | 3D Structure of DotU from Francisella novicida | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, SODIUM ION, ... | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2011-12-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Structure of the Conserved Type Six Secretion Protein Tssl (Dotu) from Francisella Novicida
J.Mol.Biol., 419, 2012
|
|
4ACK
 
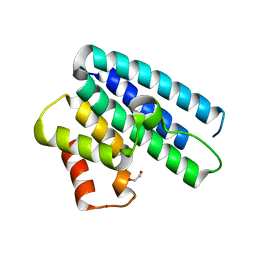 | | 3D Structure of DotU from Francisella novicida | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, TSSL | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of the Conserved Type Six Secretion Protein Tssl (Dotu) from Francisella Novicida
J.Mol.Biol., 419, 2012
|
|
5AKO
 
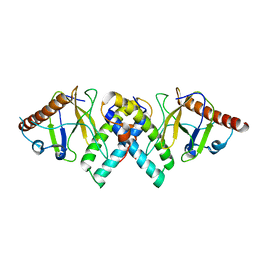 | |
6G5Q
 
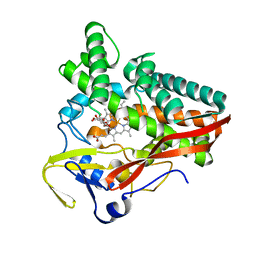 | | The structure of a carbohydrate active P450 | | Descriptor: | 6-O-methyl-beta-D-galactopyranose, Cytochrome P450, GLYCEROL, ... | | Authors: | Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Specificity and mechanism of carbohydrate demethylation by cytochrome P450 monooxygenases.
Biochem. J., 475, 2018
|
|
6Q75
 
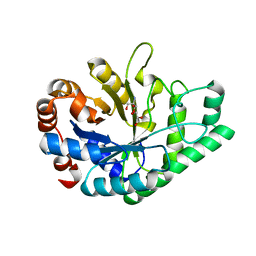 | |
6G5O
 
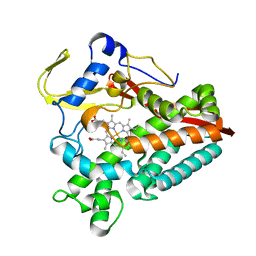 | | The structure of a carbohydrate active P450 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specificity and mechanism of carbohydrate demethylation by cytochrome P450 monooxygenases.
Biochem. J., 475, 2018
|
|
6Q78
 
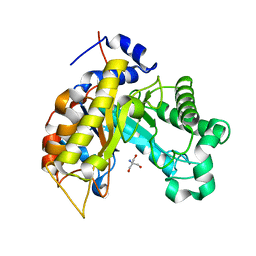 | |
4B62
 
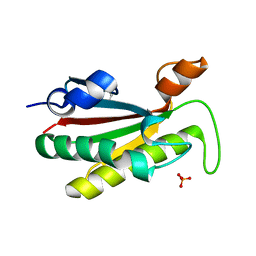 | | The structure of the cell wall anchor of the T6SS from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, TSSL1 | | Authors: | Robb, C.S, Carlson, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Periplasmic Peptidoglycan Binding Anchor of a T6Ss from Pseudomonas Aeruginosa.
To be Published
|
|
4C18
 
 | |
6HPD
 
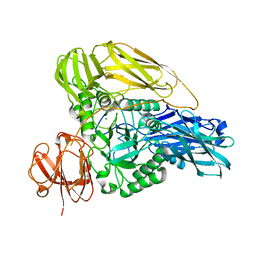 | | The structure of a beta-glucuronidase from glycoside hydrolase family 2 | | Descriptor: | BROMIDE ION, Beta-galactosidase (GH2), MAGNESIUM ION | | Authors: | Robb, C.S, Gerlach, N, Reisky, L, Bornshoeru, U, Hehemann, J.H. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
5NGW
 
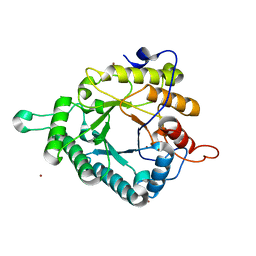 | | Glycoside hydrolase-like protein | | Descriptor: | BROMIDE ION, OpGH99A | | Authors: | Robb, C.S, Hehemann, J.-H. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a marine glycoside hydrolase family 99-related protein lacking catalytic machinery.
Protein Sci., 26, 2017
|
|
7LHA
 
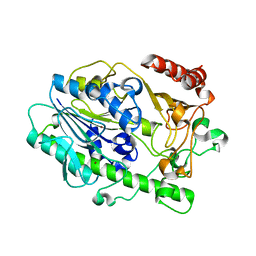 | |
7LJJ
 
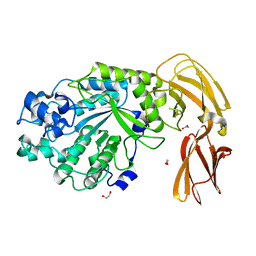 | |
7LJ2
 
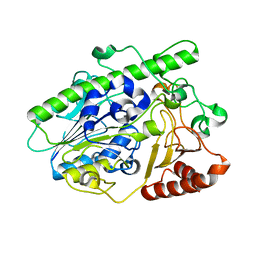 | |
7LH6
 
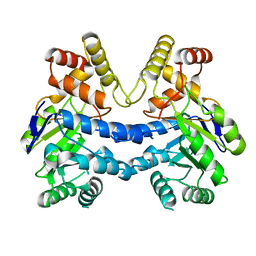 | |
5AMU
 
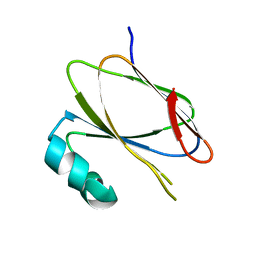 | | IglE I39A,Y40A,V44A | | Descriptor: | IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure, Dimerisation and Impact on Intramacrophage Replication of Igle in Francisella Novicida
To be Published
|
|
5AMT
 
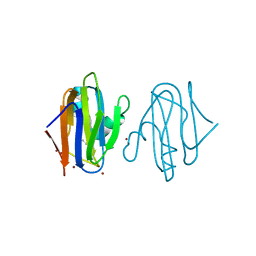 | | Intracellular growth locus protein E | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Intracellular Growth Locus E (Igle) Protein from Francisella Tularensis Subsp. Novicida.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6XFL
 
 | | Structural characterization of the type III secretion system pilotin-secretin complex InvH-InvG by NMR spectroscopy | | Descriptor: | Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFK
 
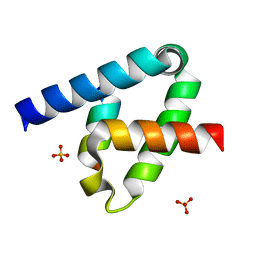 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | Descriptor: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6GCZ
 
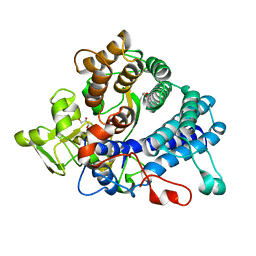 | | Laminarin binding SusD-like protein | | Descriptor: | GLYCEROL, Starch-binding associating with outer membrane | | Authors: | Mystowka, A, Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes.
FEBS J., 285, 2018
|
|
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
5A6K
 
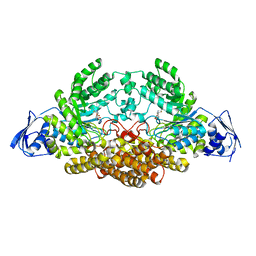 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-NGT | | Descriptor: | (2S,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, GH20C | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Second Beta-Hexosaminidase Encoded in the Streptococcus Pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
J.Biol.Chem., 290, 2015
|
|
5AC5
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
6XFJ
 
 | | Crystal structure of the type III secretion pilotin InvH | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
