5OK0
 
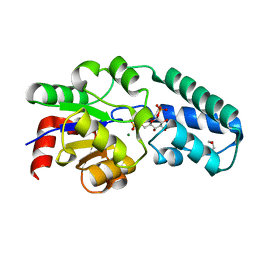 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
5I9P
 
 | |
5IGE
 
 | |
6HDI
 
 | |
5OK2
 
 | |
5OK1
 
 | |
5IGF
 
 | |
6H8V
 
 | | Beta-phosphoglucomutase from Lactococcus lactis in an open conformer in the P21 spacegroup to 1.8 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6H94
 
 | | T16A variant of beta-phosphoglucomutase from Lactococcus lactis with phosphate and TRIS bound in an open conformer to 1.5 A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
4ZIF
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIH
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
6H8Y
 
 | |
6H8U
 
 | |
6H8W
 
 | |
6H92
 
 | |
4GX8
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
6I03
 
 | |
6H8Z
 
 | |
6H91
 
 | |
4MT2
 
 | |
4BZU
 
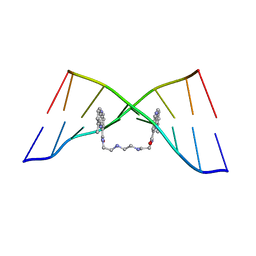 | | The Solution Structure of the MLN 944-d(TATGCATA)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4HTG
 
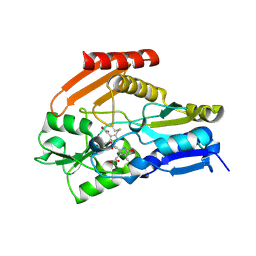 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BZV
 
 | | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
6HDJ
 
 | |
