5CV4
 
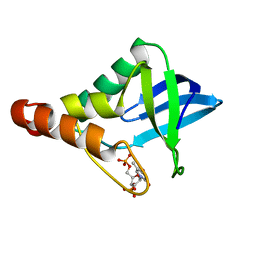 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/V66H at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Theodoru, A, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/V66H at cryogenic temperature
To be Published
|
|
5CV6
 
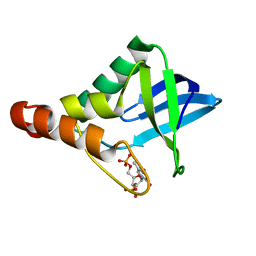 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/I92E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Theodoru, A, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/I92E at cryogenic temperature
To be Published
|
|
2R5P
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Indinavir | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, ... | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
3QGK
 
 | |
6HDL
 
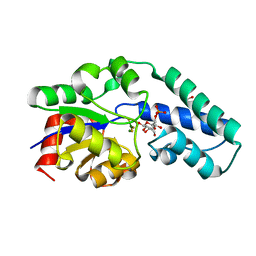 | | R49K variant of beta-phosphoglucomutase from Lactococcus lactis complexed with magnesium trifluoride and beta-G6P to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6QYK
 
 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYN
 
 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYO
 
 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5CV5
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K64G/V66K/E67G at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Ortega Quintanilla, G, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS K64G/V66K/E67G at cryogenic temperature
To be Published
|
|
3QGA
 
 | |
2R5Q
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
2R8N
 
 | | Structural Analysis of the Unbound Form of HIV-1 Subtype C Protease | | Descriptor: | GLYCEROL, Pol protein | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-11 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of unbound human immunodeficiency virus 1 subtype C protease: implications of flap dynamics and drug resistance.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2R9B
 
 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
3RGE
 
 | |
1R70
 
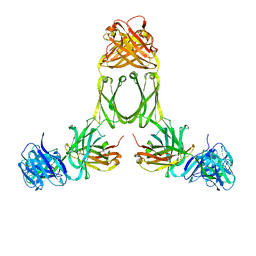 | | Model of human IgA2 determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Human IgA2(m1) Heavy Chain, Human IgA2(m1) Light Chain | | Authors: | Furtado, P.B, Whitty, P.W, Robertson, A, Eaton, J.T, Almogren, A, Kerr, M.A, Woof, J.M, Perkins, S.J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
J.Mol.Biol., 338, 2004
|
|
3SNI
 
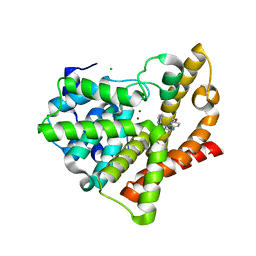 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
6QYL
 
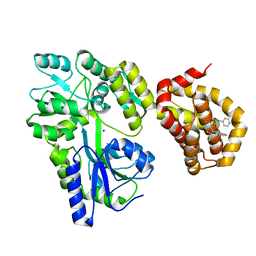 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
3PWE
 
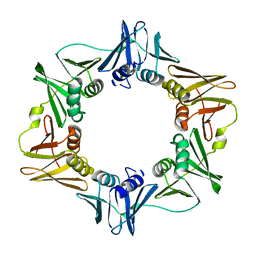 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
6QZ8
 
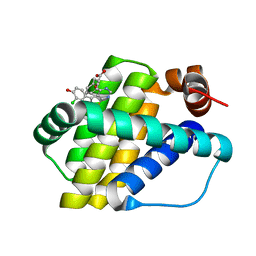 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2WN5
 
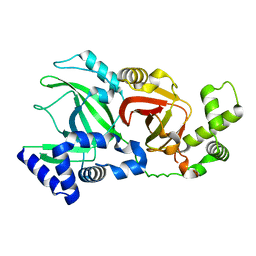 | |
6QYP
 
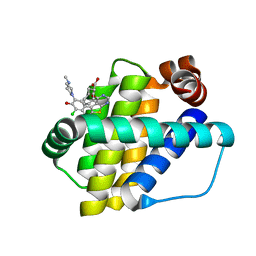 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ6
 
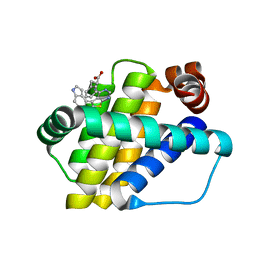 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2WN7
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2WN8
 
 | |
