3TWH
 
 | | Selenium Derivatized RNA/DNA Hybrid in complex with RNase H Catalytic Domain D132N Mutant | | Descriptor: | DNA (5'-D(*AP*TP*(SDG)P*TP*CP*(SDG))-3'), MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rob, A, Gerlits, O, Jiang, J.S, Gan, J.H, Huang, Z. | | Deposit date: | 2011-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Novel complex MAD phasing and RNase H structural insights using selenium oligonucleotides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5W63
 
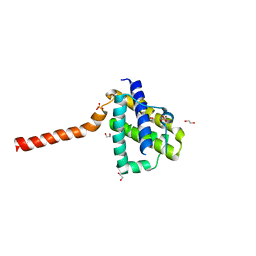 | | Crystal structure of channel catfish BAX | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator bax, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
6DE9
 
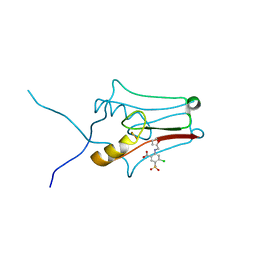 | | mitoNEET bound to furosemide | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Robart, A.R, Geldenhuys, W.J. | | Deposit date: | 2018-05-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand.
Commun Chem, 2, 2019
|
|
5W5X
 
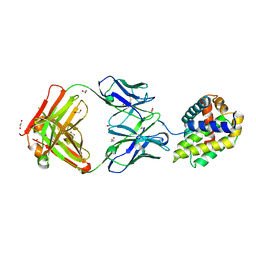 | | Crystal structure of BAXP168G in complex with an activating antibody | | Descriptor: | 1,2-ETHANEDIOL, 3C10 Fab' heavy chain, 3C10 Fab' light chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
4EQO
 
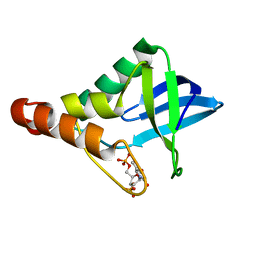 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V99D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V99D at cryogenic temperature
To be Published
|
|
5W62
 
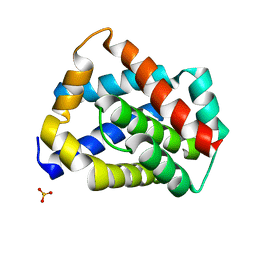 | | Crystal structure of mouse BAX monomer | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5W5Z
 
 | | Crystal structure of BAXP168G in complex with an activating antibody at high resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C10 Fab heavy chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5W60
 
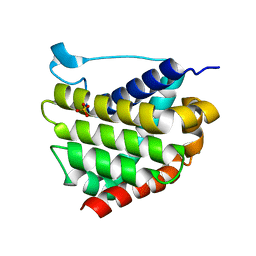 | |
4EQN
 
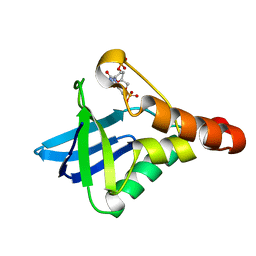 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature
To be Published
|
|
5ICG
 
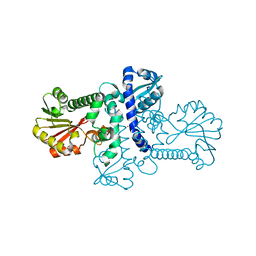 | | Crystal structure of apo (S)-norcoclaurine 6-O-methyltransferase | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, POTASSIUM ION | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5W61
 
 | |
6ZGE
 
 | | Uncleavable Spike Protein of SARS-CoV-2 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZGG
 
 | |
2Q9L
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ctag-iMazG (P43212) | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
2Q5Z
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ntag-iMazG (P43212) | | Descriptor: | GLYCEROL, Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
2Q73
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ctag-iMazG (P41212) | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
5ACN
 
 | |
4ZIF
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
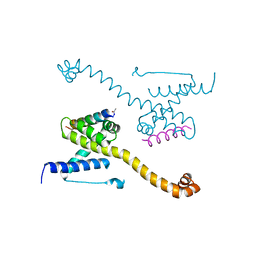 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
6EB6
 
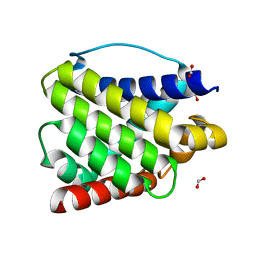 | | Crystal structure of BAX W139A monomer | | Descriptor: | Apoptosis regulator BAX, FORMIC ACID | | Authors: | Robin, A.Y. | | Deposit date: | 2018-08-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | BAX Activation: Mutations Near Its Proposed Non-canonical BH3 Binding Site Reveal Allosteric Changes Controlling Mitochondrial Association.
Cell Rep, 27, 2019
|
|
5ICC
 
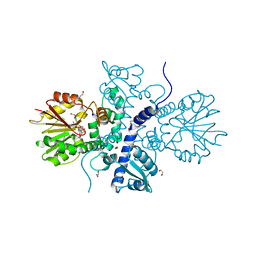 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, POTASSIUM ION, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5ICF
 
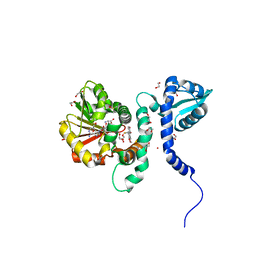 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
7BBH
 
 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
5ICE
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and norlaudanosoline | | Descriptor: | (1S)-1-(3,4-dihydroxybenzyl)-1,2,3,4-tetrahydroisoquinoline-6,7-diol, (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
6ZGF
 
 | | Spike Protein of RaTG13 Bat Coronavirus in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
