5L0R
 
 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY3
 
 | |
5KY7
 
 | | mouse POFUT1 in complex with O-glucosylated EGF(+) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EGF(+), GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
7KLG
 
 | | SARS-CoV-2 RBD in complex with Fab 15033 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033 heavy chain, Fab 15033 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KML
 
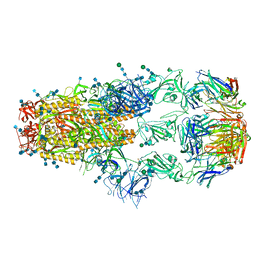 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLH
 
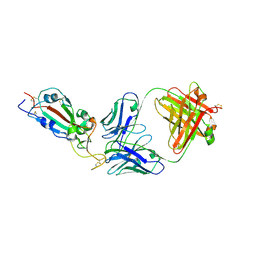 | | SARS-CoV-2 RBD in complex with Fab 15033-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, Fab 15033-7 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KMK
 
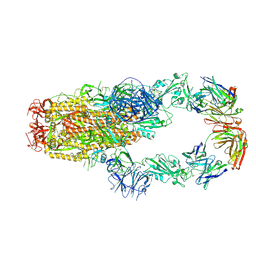 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
1GGI
 
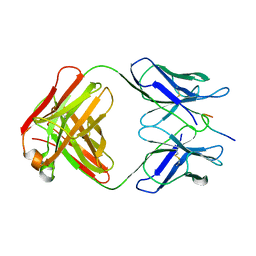 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1FOA
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FO8
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, METHYL MERCURY ION | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FO9
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1EDH
 
 | | E-CADHERIN DOMAINS 1 AND 2 IN COMPLEX WITH CALCIUM | | Descriptor: | CALCIUM ION, E-CADHERIN, MERCURY (II) ION | | Authors: | Nagar, B, Overduin, M, Ikura, M, Rini, J.M. | | Deposit date: | 1996-05-15 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of calcium-induced E-cadherin rigidification and dimerization.
Nature, 380, 1996
|
|
3M2M
 
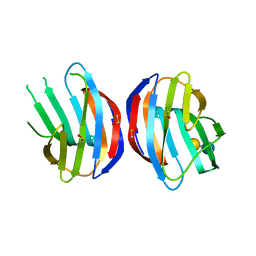 | |
3OTK
 
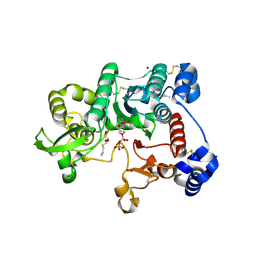 | | Structure and mechanisim of core 2 beta1,6-n-acetylglucosaminyltransferase: a Metal-ion independent gt-a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase, ... | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic characterization of leukocyte-type core 2 beta 1,6-N-acetylglucosaminyltransferase: a metal-ion-independent GT-A glycosyltransferase.
J.Mol.Biol., 414, 2011
|
|
1A3K
 
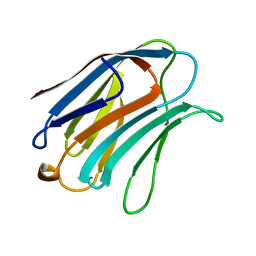 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
3BGF
 
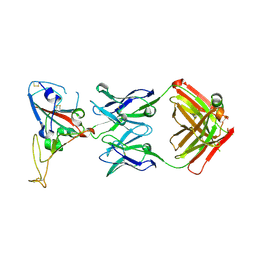 | |
1HLC
 
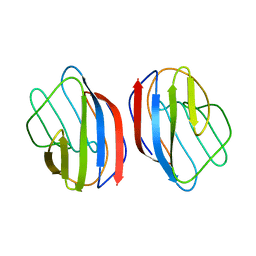 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN DIMERIC S-LAC LECTIN, L-14-II, IN COMPLEX WITH LACTOSE AT 2.9 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN LECTIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Lobsanov, Y.D, Gitt, M.A, Leffler, H, Barondes, S, Rini, J.M. | | Deposit date: | 1993-10-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution.
J.Biol.Chem., 268, 1993
|
|
4FYQ
 
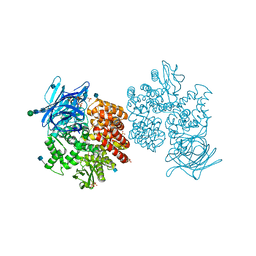 | | Human aminopeptidase N (CD13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
4FYS
 
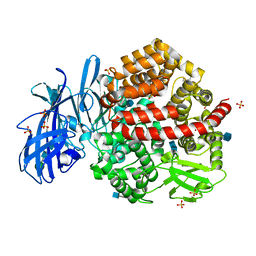 | | Human aminopeptidase N (CD13) in complex with angiotensin IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
4FYT
 
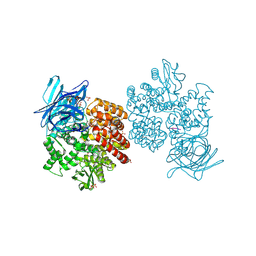 | | Human aminopeptidase N (CD13) in complex with amastatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMASTATIN, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
4FYR
 
 | | Human aminopeptidase N (CD13) in complex with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
1C3D
 
 | | X-RAY CRYSTAL STRUCTURE OF C3D: A C3 FRAGMENT AND LIGAND FOR COMPLEMENT RECEPTOR 2 | | Descriptor: | C3D, GLYCEROL | | Authors: | Nagar, B, Jones, R.G, Diefenbach, R.J, Isenman, D.E, Rini, J.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2.
Science, 280, 1998
|
|
1MUQ
 
 | | X-ray Crystal Structure of Rattlesnake Venom Complexed With Thiodigalactoside | | Descriptor: | 1-thio-beta-D-galactopyranose-(1-1)-beta-D-galactopyranose, CALCIUM ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2002-09-24 | | Release date: | 2003-07-01 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
2GAK
 
 | |
1KJL
 
 | | High Resolution X-Ray Structure of Human Galectin-3 in complex with LacNAc | | Descriptor: | BROMIDE ION, CHLORIDE ION, Galectin-3, ... | | Authors: | Sorme, P, Arnoux, P, Kahl-Knutsson, B, Leffler, H, Rini, J.M, Nilsson, U.J. | | Deposit date: | 2001-12-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and thermodynamic studies on cation-Pi interactions in lectin-ligand complexes: high-affinity galectin-3 inhibitors through fine-tuning of an arginine-arene interaction.
J.Am.Chem.Soc., 127, 2005
|
|
