1ENO
 
 | |
1ENP
 
 | |
1DQU
 
 | | CRYSTAL STRUCTURE OF THE ISOCITRATE LYASE FROM ASPERGILLUS NIDULANS | | Descriptor: | ISOCITRATE LYASE | | Authors: | Britton, K.L, Langridge, S.J, Baker, P.J, Weeradechapon, K, Sedelnikova, S.E, De Lucas, J.R, Rice, D.W, Turner, G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans.
Structure Fold.Des., 8, 2000
|
|
1DFI
 
 | |
1DFH
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND THIENO-DIAZABORINE | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
1F8G
 
 | | THE X-RAY STRUCTURE OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE FROM RHODOSPIRILLUM RUBRUM COMPLEXED WITH NAD+ | | Descriptor: | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Buckley, P.A, Baz Jackson, J, Schneider, T, White, S.A, Rice, D.W, Baker, P.J. | | Deposit date: | 2000-06-30 | | Release date: | 2001-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Structure Fold.Des., 8, 2000
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
1D8A
 
 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
1CUK
 
 | |
1DFG
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND BENZO-DIAZABORINE | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
1CWU
 
 | | BRASSICA NAPUS ENOYL ACP REDUCTASE A138G MUTANT COMPLEXED WITH NAD+ AND THIENODIAZABORINE | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ENOYL ACP REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Roujeinikova, A, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor binding studies on enoyl reductase reveal conformational changes related to substrate recognition.
J.Biol.Chem., 274, 1999
|
|
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1GJW
 
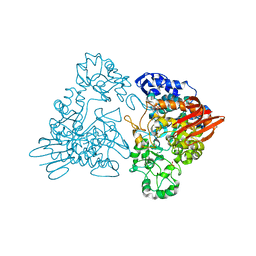 | | Thermotoga maritima maltosyltransferase complex with maltose | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
1G2A
 
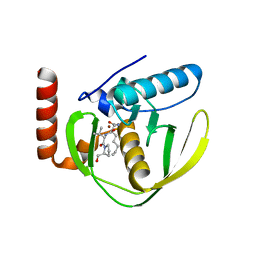 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1GJU
 
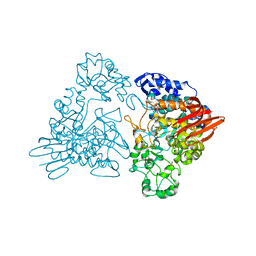 | | Maltosyltransferase from Thermotoga maritima | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-02 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
