9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis
Nature Communications, 2024
|
|
7ZH7
 
 | |
6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | Descriptor: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | Authors: | Costanzi, E, Sormanni, P, Ricagno, S. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
3NA4
 
 | | D53P beta-2 microglobulin mutant | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Beta-2-microglobulin | | Authors: | Azinas, S, Ricagno, S, Bolognesi, M. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-strand perturbation and amyloid propensity in beta-2 microglobulin
Febs J., 278, 2011
|
|
3IB4
 
 | |
7P0T
 
 | |
7P0A
 
 | |
8P89
 
 | |
8P88
 
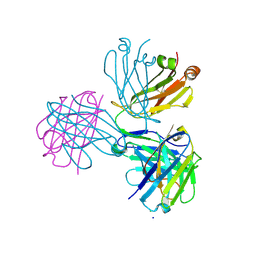 | |
8QOS
 
 | | Capra hircus reactive intermediate deaminase A - V25W | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, POTASSIUM ION | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Capra hircus reactive intermediate deaminase A - V25W
To Be Published
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
1GTH
 
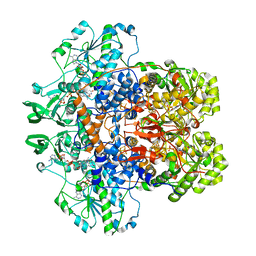 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GT8
 
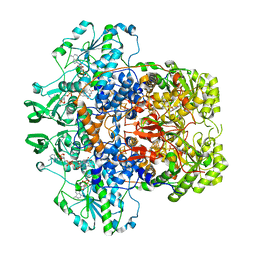 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
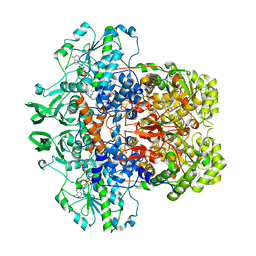 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
8Q3H
 
 | | Capra hircus reactive intermediate deaminase A mutant - A108D | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, GLYCEROL | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capra hircus reactive intermediate deaminase A mutant - A108D
To Be Published
|
|
8QOK
 
 | |
8QOP
 
 | |
8QOQ
 
 | |
8QOM
 
 | |
8QOV
 
 | | Capra hircus reactive intermediate deaminase A mutant - I126Y | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Capra hircus reactive intermediate deaminase A mutant - I126Y
To Be Published
|
|
8QOU
 
 | | Reactive intermediate deaminase A mutant - R107K | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reactive intermediate deaminase A mutant - R107K
To Be Published
|
|
3TLR
 
 | |
3TM6
 
 | |
