8S8W
 
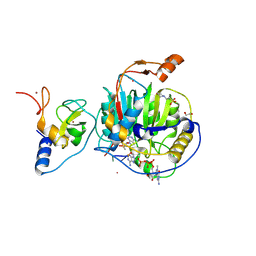 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
8S8X
 
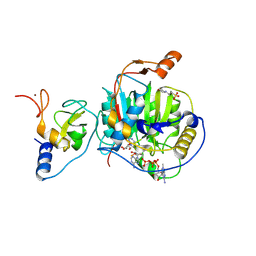 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
7RFO
 
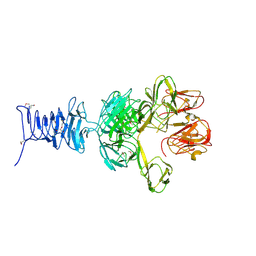 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7REJ
 
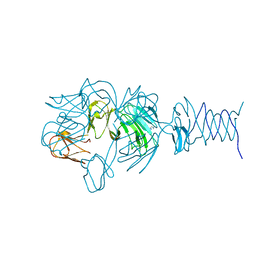 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | Descriptor: | IMIDAZOLE, Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-13 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFV
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
4A39
 
 | | Metallo-carboxypeptidase from Pseudomonas Aeruginosa in complex with (2-guanidinoethylmercapto)succinic acid | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4A38
 
 | | METALLO-CARBOXYPEPTIDASE FROM PSEUDOMONAS AUREGINOSA IN COMPLEX WITH L-BENZYLSUCCINIC ACID | | Descriptor: | L-BENZYLSUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4YPS
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4A37
 
 | | Metallo-carboxypeptidase from Pseudomonas Aeruginosa | | Descriptor: | METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
1A6G
 
 | | CARBONMONOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-25 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6K
 
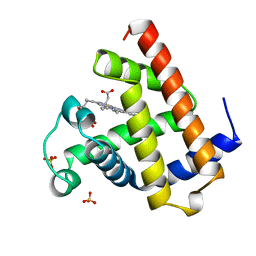 | | AQUOMET-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Berendzen, J, Chu, K, Schlichting, I, Sweet, R.M. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6M
 
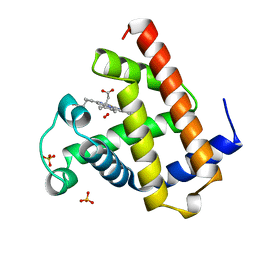 | | OXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6N
 
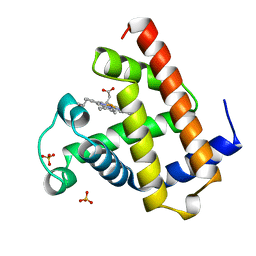 | | DEOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
3P2P
 
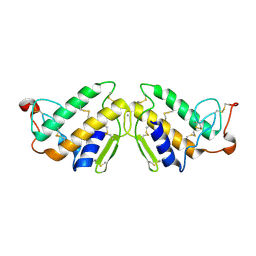 | | ENHANCED ACTIVITY AND ALTERED SPECIFICITY OF PHOSPHOLIPASE A2 BY DELETION OF A SURFACE LOOP | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1989-11-29 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced activity and altered specificity of phospholipase A2 by deletion of a surface loop.
Science, 244, 1989
|
|
4ZZN
 
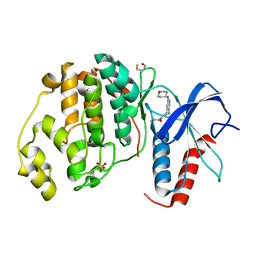 | | Human ERK2 in complex with an inhibitor | | Descriptor: | 2-[[5-chloranyl-2-(oxan-4-ylamino)pyridin-4-yl]amino]-N-methyl-benzamide, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
4ZZM
 
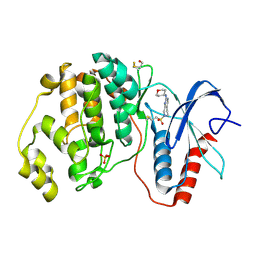 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | 7-ethylsulfonyl-N-(oxan-4-yl)-6,8-dihydro-5H-pyrido[3,4-d]pyrimidin-2-amine, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
1JE4
 
 | |
3WOV
 
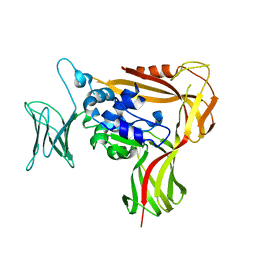 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PaAglB-L, Q9V250_PYRAB, PAB2202) from Pyrococcus abyssi | | Descriptor: | CALCIUM ION, Oligosaccharyl transferase | | Authors: | Matsuoka, R, Nyirenda, J, Maita, N, Kohda, D. | | Deposit date: | 2014-01-05 | | Release date: | 2014-01-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PaAglB-L, Q9V250_PYRAB, PAB2202) from Pyrococcus abyssi
To be Published
|
|
5JO3
 
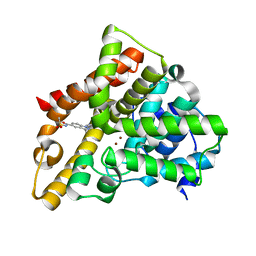 | | PDE5A for NaV1.7 | | Descriptor: | 1-(5-chloro-6-methoxypyridin-3-yl)-3-methyl-N-(methylsulfonyl)-1H-indazole-5-carboxamide, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Storer, I, Chrencik, J. | | Deposit date: | 2016-05-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PDE5A for NaV1.7
To be published
|
|
8VLL
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLK
 
 | | Crystal structure of the yeast cytosine deaminase containing both open and closed active sites | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, SULFATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
