4WYU
 
 | |
4WYT
 
 | |
5ZBS
 
 | | Crystal Structure of Kinesin-3 KIF13B motor Y73C mutant | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
5ZBR
 
 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
6A1Z
 
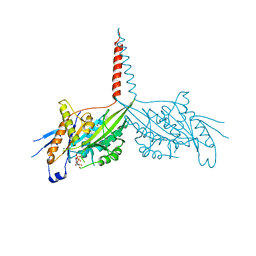 | | Crystal Structure of dimeric Kinesin-3 KIF13B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin family member 13B, MAGNESIUM ION | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-06-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A20
 
 | | Crystal Structure of auto-inhibited Kinesin-3 KIF13B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXAETHYLENE GLYCOL, Kinesin family member 13B, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-06-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7WEK
 
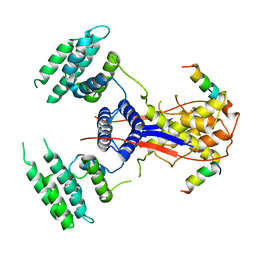 | |
7WEJ
 
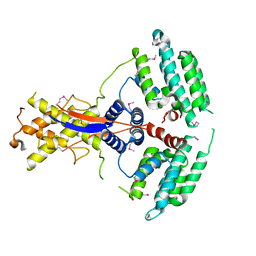 | | Crystal structure of the mouse Wdr47 NTD. | | Descriptor: | WD repeat-containing protein 47 | | Authors: | Ren, J.Q, Li, D, Feng, W. | | Deposit date: | 2021-12-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Intertwined Wdr47-NTD dimer recognizes a basic-helical motif in Camsap proteins for proper central-pair microtubule formation.
Cell Rep, 41, 2022
|
|
6KLR
 
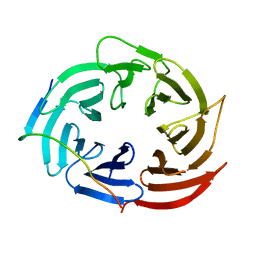 | |
5DJO
 
 | | Crystal structure of the CC1-FHA tandem of Kinesin-3 KIF13A | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Ren, J.Q, Li, W, Huo, L, Feng, W. | | Deposit date: | 2015-09-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
J.Biol.Chem., 291, 2016
|
|
5DJN
 
 | |
7D8V
 
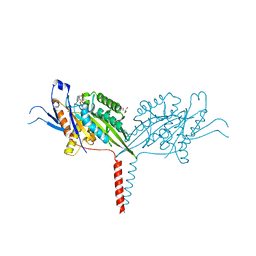 | | Crystal Structure of A Kinesin-3 KIF13B mutant-T192Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin family member 13B, ... | | Authors: | Ren, J.Q, Feng, W. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motor domain-mediated autoinhibition dictates axonal transport by the kinesin UNC-104/KIF1A.
Plos Genet., 17, 2021
|
|
5HPY
 
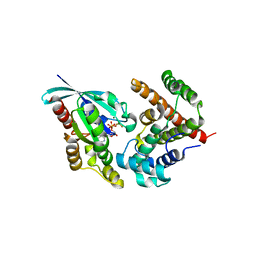 | | Crystal Structure of RhoA.GDP.MgF3-in complex with human Myosin 9b RhoGAP domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRIFLUOROMAGNESATE, ... | | Authors: | Yi, F.S, Ren, J.Q, Feng, W. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncanonical Myo9b-RhoGAP Accelerates RhoA GTP Hydrolysis by a Dual-Arginine-Finger Mechanism
J.Mol.Biol., 428, 2016
|
|
6A5Q
 
 | |
6A5S
 
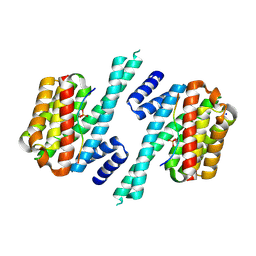 | | Structure of 14-3-3 gamma in complex with TFEB 14-3-3 binding motif | | Descriptor: | 14-3-3 protein gamma, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Xu, Y, Ren, J.Q, Feng, W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization.
Autophagy, 15, 2019
|
|
7WRG
 
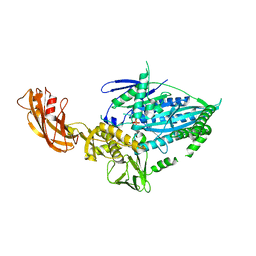 | | Crystal structure of full-length kinesin-3 KLP-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein, MAGNESIUM ION | | Authors: | Wang, W.J, Ren, J.Q, Song, W.Y, Feng, W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition.
Nat Commun, 13, 2022
|
|
7XKC
 
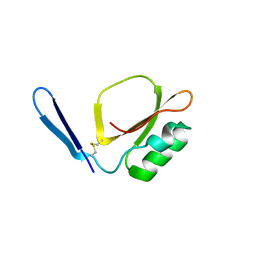 | | Crystal structure of Daucus Carrot hypoglycemic peptide (DCHP) | | Descriptor: | DCHP, SULFATE ION | | Authors: | Guo, T, Ren, J.Q, Wang, L, Shi, Y.W, Feng, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Characterization of a thermostable, protease-tolerant inhibitor of alpha-glycosidase from carrot: A potential oral additive for treatment of diabetes.
Int.J.Biol.Macromol., 209, 2022
|
|
6IYY
 
 | |
5ED9
 
 | | Crystal structure of CC1 of mouse SUN2 | | Descriptor: | SUN domain-containing protein 2 | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5ED8
 
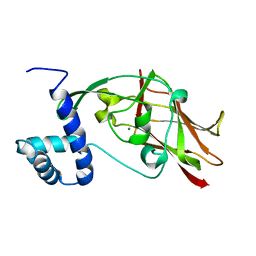 | | Crystal structure of CC2-SUN of mouse SUN2 | | Descriptor: | MAGNESIUM ION, MKIAA0668 protein | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5C5S
 
 | |
5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5E6N
 
 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
