4ZUJ
 
 | |
4ZQ3
 
 | |
4ZUI
 
 | |
6BSY
 
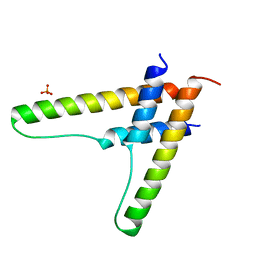 | | HIV-1 Rev assembly domain (residues 1-69) | | Descriptor: | PHOSPHATE ION, Protein Rev | | Authors: | Watts, N.R, Eren, E, Zhuang, X, Wang, Y.X, Steven, A.C, Wingfield, P.T. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export.
J. Struct. Biol., 203, 2018
|
|
7SVS
 
 | |
6CYX
 
 | |
6CYL
 
 | |
6G8R
 
 | | SP140 PHD-Bromodomain complex with scFv | | Descriptor: | 1,2-ETHANEDIOL, Nuclear body protein SP140, ZINC ION, ... | | Authors: | Fairhead, M, Graslund, S, Strain-Damerell, C, Picaud, S.S, Pike, A.C.W, Pinkas, D.M, Wigren, E, Preger, C, Persson Lotsholm, H, Ossipova, E, Filippakopoulos, P, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | SP140 PHD-Bromodomain complex with scFv
To Be Published
|
|
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHN
 
 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | Descriptor: | CHLORIDE ION, Thaumatin-1 | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHB
 
 | | 3D electron diffraction structure of bovine insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6CYP
 
 | |
6CYS
 
 | |
4KHV
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23S at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23S at cryogenic temperature
TO BE PUBLISHED
|
|
4NKL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A/I92Q at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-12 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4NMZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23Q/V66A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities in proteins
To be Published
|
|
4K6D
 
 | | Crystal structure of Staphylococcal nuclease variant V23M/T62F at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-15 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure effects on proteins
To be Published
|
|
4K5X
 
 | | Crystal structure of Staphylococcal nuclease variant V23M/L36F at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-15 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure effects on proteins
To be Published
|
|
4KD4
 
 | | Crystal structure of Staphylococcal nuclease variant V23L/L25V/V66I/I72V at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
5EKK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D/L125E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Siegler, M.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D/L125E at cryogenic temperature
To be Published
|
|
5EKL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62D/N100E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62D/N100E at cryogenic temperature
To be Published
|
|
5MJ4
 
 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 2 | | Descriptor: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Henderikx, P, Lasters, I, Savvides, S. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | STRUCTURAL BASIS OF IL-23 ANTAGONISM BY AN ALPHABODY PROTEIN
Nature Communications, 5, 2014
|
|
5F2D
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N/V99T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N/V99T at cryogenic temperature
To be Published
|
|
8QEY
 
 | | Structure of human Asc1/CD98hc heteromeric amino acid transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Martinez-Molledo, M, Rullo-Tubau, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2023-09-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and mechanisms of transport of human Asc1/CD98hc amino acid transporter.
Nat Commun, 15, 2024
|
|
5MJ3
 
 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 1 | | Descriptor: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Hendrikx, P, Lasters, I, Savvides, S.N. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis Of Il-23 Antagonism By An Alphabody Protein Scaffold.
Nat Commun, 5, 2014
|
|
