6V1R
 
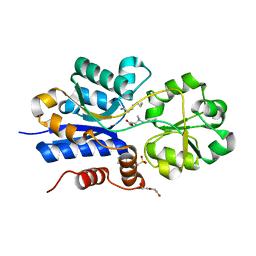 | | Crystal structure of iAChSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETYLCHOLINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A genetically encoded fluorescent sensor for in vivo acetylcholine detection
To Be Published
|
|
6VQU
 
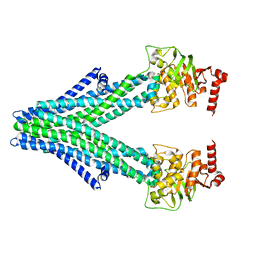 | | Structure of a bacterial Atm1-family ABC exporter | | Descriptor: | ATM1-type heavy metal exporter | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A structural framework for unidirectional transport by a bacterial ABC exporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQT
 
 | |
6WA8
 
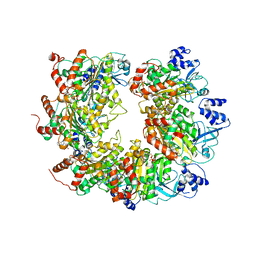 | |
2CXB
 
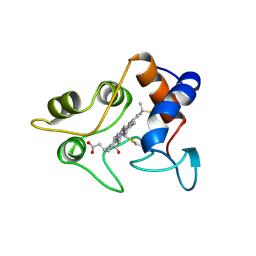 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-04-21 | | Release date: | 1995-10-15 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PSS
 
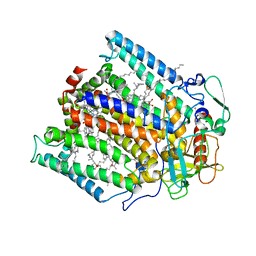 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
1PST
 
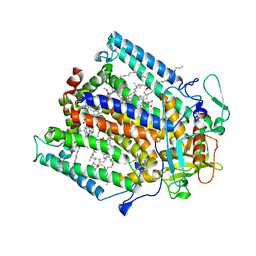 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
7S7Y
 
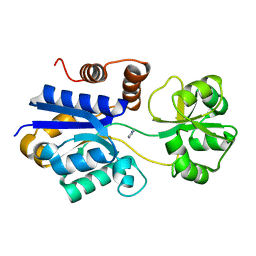 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein | | Descriptor: | IMIDAZOLE, iNicSnFR 4.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S7Z
 
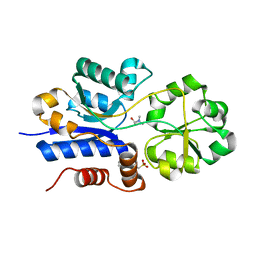 | | Crystal structure of iAChSnFR Acetylcholine Sensor precursor binding protein with choline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHOLINE ION, iAchSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S7W
 
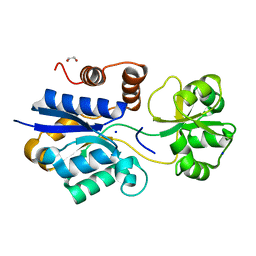 | | Crystal structure of iNicSnFR3a Nicotine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, iNicSnFR 3.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S80
 
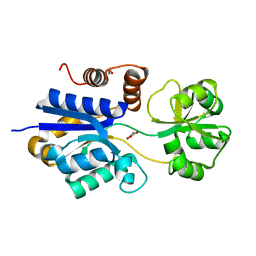 | | Crystal structure of iAChSnFR Acetylcholine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, MALONATE ION, iAchSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S7X
 
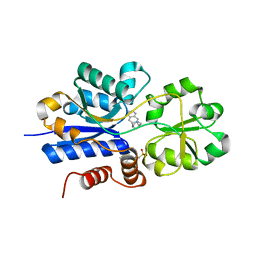 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein with varenicline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, VARENICLINE, ... | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
1AFC
 
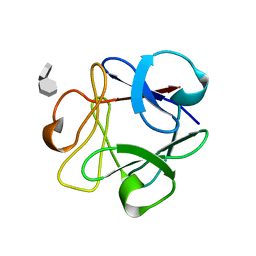 | | STRUCTURAL STUDIES OF THE BINDING OF THE ANTI-ULCER DRUG SUCROSE OCTASULFATE TO ACIDIC FIBROBLAST GROWTH FACTOR | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Hsu, B.T, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the binding of the anti-ulcer drug sucrose octasulfate to acidic fibroblast growth factor.
Structure, 1, 1993
|
|
1BFC
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN HEXAMER FRAGMENT | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Faham, S, Rees, D.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
1WOD
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE PROTEIN, COMPLEXED WITH TUNGSTATE | | Descriptor: | MODA, TUNGSTATE(VI)ION | | Authors: | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
1BFB
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN TETRAMER FRAGMENT | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Faham, S, Rees, D.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
1AOR
 
 | | STRUCTURE OF A HYPERTHERMOPHILIC TUNGSTOPTERIN ENZYME, ALDEHYDE FERREDOXIN OXIDOREDUCTASE | | Descriptor: | ALDEHYDE FERREDOXIN OXIDOREDUCTASE, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Chan, M.K, Mukund, S, Kletzin, A, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1995-02-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase.
Science, 267, 1995
|
|
2AFI
 
 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
2AFH
 
 | | Crystal Structure of Nucleotide-Free Av2-Av1 Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
3DHX
 
 | | Crystal structure of isolated C2 domain of the methionine uptake transporter | | Descriptor: | IODIDE ION, Methionine import ATP-binding protein metN | | Authors: | Johnson, E, Kaiser, J.T, Lee, A.T, Rees, D.C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
2NIP
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
2NQ2
 
 | | An inward-facing conformation of a putative metal-chelate type ABC transporter. | | Descriptor: | Hypothetical ABC transporter ATP-binding protein HI1470, Hypothetical ABC transporter permease protein HI1471 | | Authors: | Pinkett, H.P, Lee, A.T, Lum, P, Locher, K.P, Rees, D.C. | | Deposit date: | 2006-10-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An inward-facing conformation of a putative metal-chelate-type ABC transporter.
Science, 315, 2007
|
|
7T4H
 
 | | Selenium-incorporated nitrogenase Fe protein (Av2-Se) from A. vinelandii (22 mM KSeCN, with Av1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fe4-Se4 cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Buscagan, T.M, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2021-12-09 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selenocyanate derived Se-incorporation into the Nitrogenase Fe protein cluster.
Elife, 11, 2022
|
|
7TPZ
 
 | | Selenium-free nitrogenase Fe protein (Av2) from A. vinelandii (nucleotide control) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Buscagan, T.M, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selenocyanate derived Se-incorporation into the Nitrogenase Fe protein cluster.
Elife, 11, 2022
|
|
7TQ0
 
 | | Selenium-incorporated nitrogenase Fe protein (Av2-Se) from A. vinelandii (22 mM KSeCN, with Av1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fe4-Se4 cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Buscagan, T.M, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Selenocyanate derived Se-incorporation into the Nitrogenase Fe protein cluster.
Elife, 11, 2022
|
|
