6G6U
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 2.7 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, NITRATE ION, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6G73
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 3.3 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
5UZ4
 
 | | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly | | Descriptor: | 16S RIBOSOMAL RNA, 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, 30S ribosomal protein S10, ... | | Authors: | Razi, A, Guarne, A, Ortega, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1OJ5
 
 | | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | | Descriptor: | IODIDE ION, SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 6, STEROID RECEPTOR COACTIVATOR 1A | | Authors: | Razeto, A, Ramakrishnan, V, Giller, K, Lakomek, N, Carlomagno, T, Griesinger, C, Lodrini, M, Litterst, C.M, Pftizner, E, Becker, S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain
J.Mol.Biol., 336, 2004
|
|
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
5JPZ
 
 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
2UUU
 
 | | alkyldihydroxyacetonephosphate synthase in P212121 | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, FLAVIN-ADENINE DINUCLEOTIDE, HEXADECAN-1-OL | | Authors: | Razeto, A, Mattiroli, F, Carpanelli, E, Aliverti, A, Pandini, V, Coda, A, Mattevi, A. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Structure, 15, 2007
|
|
2UUV
 
 | | alkyldihydroxyacetonephosphate synthase in P1 | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, FLAVIN-ADENINE DINUCLEOTIDE, HEXADECAN-1-OL | | Authors: | Razeto, A, Mattiroli, F, Carpanelli, E, Aliverti, A, Pandini, V, Coda, A, Mattevi, A. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Structure, 15, 2007
|
|
1Y9O
 
 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
1YPH
 
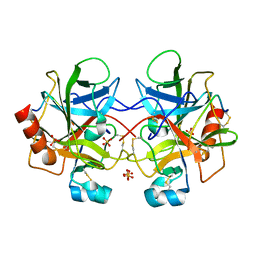 | | High resolution structure of bovine alpha-chymotrypsin | | Descriptor: | CHYMOTRYPSIN A, chain A, chain B, ... | | Authors: | Razeto, A, Galunsky, B, Kasche, V, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution structure of native bovine alpha-chymotrypsin
To be Published
|
|
1J49
 
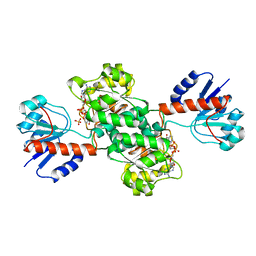 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1J4A
 
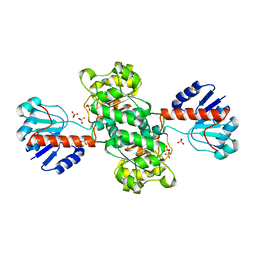 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1IRI
 
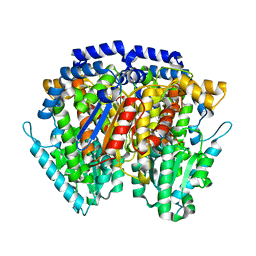 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1JIQ
 
 | | Crystal Structure of Human Autocrine Motility Factor | | Descriptor: | autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
2CXO
 
 | | Crystal structure of mouse AMF / E4P complex | | Descriptor: | D-4-PHOSPHOERYTHRONIC ACID, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXT
 
 | | Crystal structure of mouse AMF / F6P complex | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXN
 
 | | Crystal structure of mouse AMF / phosphate complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXU
 
 | | Crystal structure of mouse AMF / M6P complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXP
 
 | | Crystal structure of mouse AMF / A5P complex | | Descriptor: | ARABINOSE-5-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CVP
 
 | | Crystal structure of mouse AMF | | Descriptor: | ACETATE ION, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXQ
 
 | | Crystal structure of mouse AMF / S6P complex | | Descriptor: | D-SORBITOL-6-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXS
 
 | | Crystal structure of mouse AMF / F6P complex | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXR
 
 | | Crystal structure of mouse AMF / 6PG complex | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
6VIO
 
 | |
6MOZ
 
 | | Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor | | Descriptor: | (2R,3S,4R)-2-{[4-(3,5-dichlorophenyl)-1H-1,2,3-triazol-1-yl]methyl}pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Patterson-Orazem, A.C, Lieberman, R.L. | | Deposit date: | 2018-10-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring substituent diversity on pyrrolidine-aryltriazole iminosugars: Structural basis of beta-glucocerebrosidase inhibition.
Bioorg.Chem., 86, 2019
|
|
