5IOJ
 
 | |
1EYW
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG TRIETHYLPHOSPHATE | | Descriptor: | 2-PHENYL-ETHANOL, PHOSPHOTRIESTERASE, TRIETHYL PHOSPHATE, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Hong, S.-B. | | Deposit date: | 2000-05-09 | | Release date: | 2000-12-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The binding of substrate analogs to phosphotriesterase.
J.Biol.Chem., 275, 2000
|
|
6DXQ
 
 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
1EZ2
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIISOPROPYLMETHYL PHOSPHONATE. | | Descriptor: | METHYLPHOSPHONIC ACID DIISOPROPYL ESTER, PHOSPHOTRIESTERASE, ZINC ION | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Hong, S.-B. | | Deposit date: | 2000-05-09 | | Release date: | 2000-12-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The binding of substrate analogs to phosphotriesterase.
J.Biol.Chem., 275, 2000
|
|
2AQV
 
 | | Crystal Structure of E. coli Isoaspartyl Dipeptidase mutant Y137F | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
5HRM
 
 | |
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
1T36
 
 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
4ZSU
 
 | |
4ZST
 
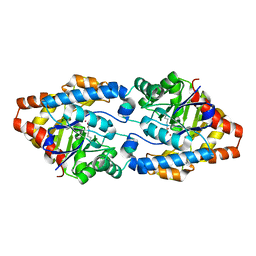 | |
1MMX
 
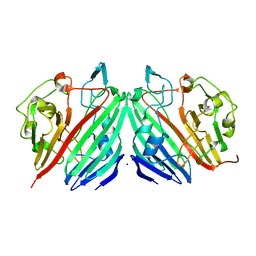 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
6DWV
 
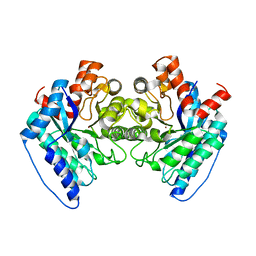 | | Crystal structure of the LigJ Hydratase in the Apo state | | Descriptor: | 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
6DXS
 
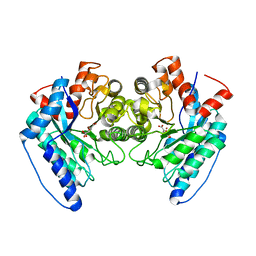 | | Crystal structure of the LigJ hydratase E284Q mutant substrate complex with (3Z)-2-keto-4-carboxy-3-hexenedioate | | Descriptor: | (2Z)-4-oxobut-2-ene-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Hogancamp, T.N. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
8V4G
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
4GLO
 
 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog
(target EFI-505321) from burkholderia multivorans, with bound NAD
To be Published
|
|
4GC3
 
 | | Crystal structure of L-HISTIDINOL PHOSPHATE PHOSPHATASE (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and sulfate | | Descriptor: | L-HISTIDINOL PHOSPHATE PHOSPHATASE, SULFATE ION, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
4GK8
 
 | | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and L-histidinol arsenate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidinol-phosphatase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
3LY0
 
 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MDW
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
6WN6
 
 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO6
 
 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
