3MDU
 
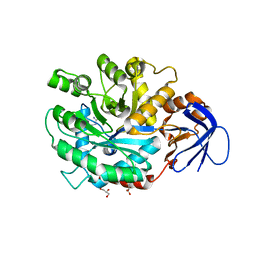 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | Descriptor: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4003 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MTW
 
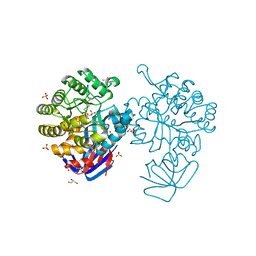 | | Crystal structure of L-Lysine, L-Arginine carboxypeptidase Cc2672 from Caulobacter Crescentus CB15 complexed with N-methyl phosphonate derivative of L-Arginine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L-Arginine carboxypeptidase Cc2672, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-05-01 | | Release date: | 2010-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Identification and Structure Determination of Two Novel Prolidases from cog1228 in the Amidohydrolase Superfamily
Biochemistry, 49, 2010
|
|
3O7U
 
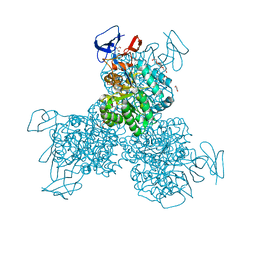 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
3N2C
 
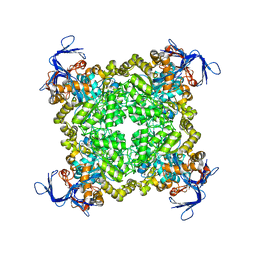 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3PNZ
 
 | | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphotriesterase family protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-11-20 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.5983 Å) | | Cite: | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes
To be Published
|
|
3GIQ
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3FDK
 
 | | Crystal structure of hydrolase DR0930 with promiscuous catalytic activity | | Descriptor: | HYDROLASE DR0930, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, L.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2008-11-25 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK5
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK8
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HKA
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Fructuronate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-fructuronic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
1JDB
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Wesenberg, G, Raushel, F.M, Rayment, I. | | Deposit date: | 1997-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1C30
 
 | | CRYSTAL STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTATION C269S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-24 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1C3O
 
 | | CRYSTAL STRUCTURE OF THE CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTANT C269S WITH BOUND GLUTAMINE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-28 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1BXR
 
 | | STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE COMPLEXED WITH THE ATP ANALOG AMPPNP | | Descriptor: | CARBAMOYL-PHOSPHATE SYNTHASE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Wesenberg, G, Raushel, F.M, Holden, H.M. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
Biochemistry, 38, 1999
|
|
1DPM
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIETHYL 4-METHYLBENZYLPHOSPHONATE | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, FORMIC ACID, PHOSPHOTRIESTERASE, ... | | Authors: | Vanhooke, J.L, Benning, M.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 1996-02-13 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
Biochemistry, 35, 1996
|
|
1CE8
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIS COLI WITH COMPLEXED WITH THE ALLOSTERIC LIGAND IMP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, INOSINIC ACID, ... | | Authors: | Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-03-18 | | Release date: | 1999-07-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase.
J.Biol.Chem., 274, 1999
|
|
1CS0
 
 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
4QS6
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID, NO METAL, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Ligw2 Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QRN
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE AND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-Carboxyvanillate Decarboxylase, ACETATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4R88
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine | | Descriptor: | 1,2-ETHANEDIOL, 5-fluorocytosine, ACETIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine
To be Published
|
|
4RDV
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
4QTG
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE | | Descriptor: | 5-CARBOXYVANILLATE DECARBOXYLASE, GLYCEROL, MANGANESE (II) ION | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase LIGW2 from Novosphingobium Aromaticivorans
To be Published
|
|
4QSF
 
 | | CRYSTAL STRUCTURE of AMIDOHYDROLASE PMI1525 (TARGET EFI-500319) FROM PROTEUS MIRABILIS HI4320, A COMPLEX WITH BUTYRIC ACID AND MANGANESE | | Descriptor: | Amidohydrolase Pmi1525, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Xiang, D.F, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
