7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
3RWP
 
 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Greasley, S.E, Hickey, M, Ferre, R.-A, Krauss, M, Cronin, C. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
4YE0
 
 | | Stress-induced protein 1 truncation mutant (43 - 140) from Caenorhabditis elegans | | Descriptor: | SULFATE ION, Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
2ACG
 
 | | ACANTHAMOEBA CASTELLANII PROFILIN II | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-08-30 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
7AT8
 
 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | Deposit date: | 2020-10-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
5MAN
 
 | |
5M9X
 
 | | Structure of sucrose phosphorylase from Bifidobacterium adolescentis bound to glycosylated resveratrol | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[3-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-5-oxidanyl-phenoxy]oxane-3,4 ,5-triol, Sucrose phosphorylase | | Authors: | Grimm, C, Kraus, M. | | Deposit date: | 2016-11-02 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Switching enzyme specificity from phosphate to resveratrol glucosylation.
Chem. Commun. (Camb.), 53, 2017
|
|
9C0Y
 
 | | Clathrin terminal domain complexed with Pitstop 2c | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
9C0Z
 
 | | Clathrin terminal domain complexed with pitstop 2d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
8FXH
 
 | |
8FXI
 
 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
5MB2
 
 | |
8EXL
 
 | | Crystal structure of PI3K-alpha in complex with taselisib | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | Descriptor: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXV
 
 | | Crystal structure of PI3K-alpha in complex with compound 32 | | Descriptor: | N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
7P5M
 
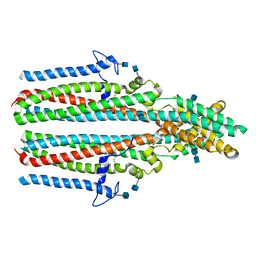 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
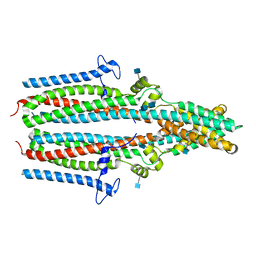 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7THX
 
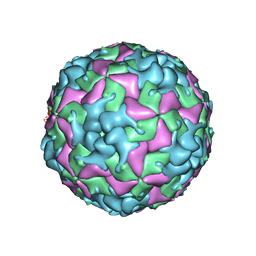 | | Cryo-EM structure of W6 possum enterovirus | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, I, Jayawardena, N, Strauss, M, Bostina, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM Structure of a Possum Enterovirus.
Viruses, 14, 2022
|
|
8CXP
 
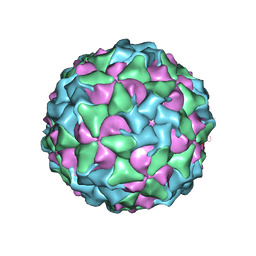 | | Characterisation of a Seneca Valley Virus Thermostable Mutant | | Descriptor: | Capsid protein VP1, Capsid protein VP3, VP2, ... | | Authors: | Jayawardena, N, Bostina, M, Strauss, M. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Characterisation of a Seneca Valley virus thermostable mutant.
Virology, 575, 2022
|
|
8UVL
 
 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
6DKG
 
 | | Crystal structure of Trk-A in complex with the Pan-Trk Kinase Inhibitor, compound 13b. | | Descriptor: | 2-{[(3R,4S)-3-fluoro-1-{[4-(trifluoromethoxy)phenyl]acetyl}piperidin-4-yl]oxy}-4-(2-hydroxy-2-methylpropoxy)benzamide, 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-05-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
J. Med. Chem., 61, 2018
|
|
