4A3V
 
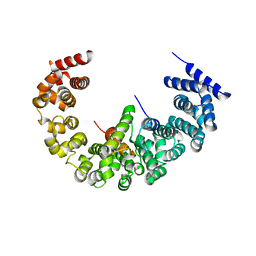 | | yeast regulatory particle proteasome assembly chaperone Hsm3 in complex with Rpt1 C-terminal fragment | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 7 HOMOLOG, DNA MISMATCH REPAIR PROTEIN HSM3, LINKER | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A3T
 
 | | yeast regulatory particle proteasome assembly chaperone Hsm3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN HSM3 | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1Z68
 
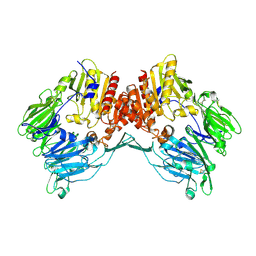 | | Crystal Structure Of Human Fibroblast Activation Protein alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aertgeerts, K, Levin, I, Shi, L, Prasad, G.S, Zhang, Y, Kraus, M.L, Salakian, S, Snell, G.P, Sridhar, V, Wijnands, R, Tennant, M.G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha.
J.Biol.Chem., 280, 2005
|
|
5MB2
 
 | |
3H1Z
 
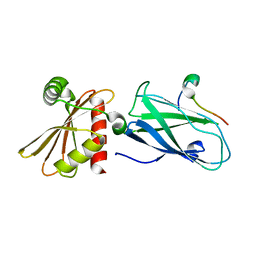 | | Molecular basis for the association of PIPKIgamma -p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit beta-1, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Krainer, G, Keller, S, Saenger, W, Krauss, M, Haucke, V. | | Deposit date: | 2009-04-14 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
3ZFS
 
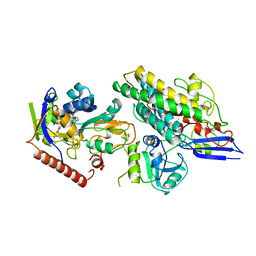 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
1RM8
 
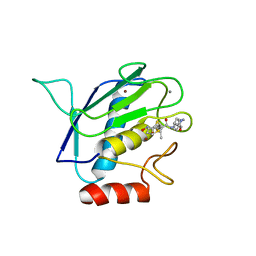 | | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: Characterization of MT-MMP specific features | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, Matrix metalloproteinase-16, ... | | Authors: | Lang, R, Braun, M, Sounni, N.E, Noel, A, Frankenne, F, Foidart, J.-M, Bode, W, Maskos, K. | | Deposit date: | 2003-11-27 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: characterization of MT-MMP specific features.
J.Mol.Biol., 336, 2004
|
|
2ACG
 
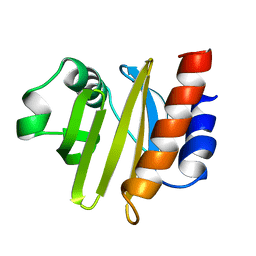 | | ACANTHAMOEBA CASTELLANII PROFILIN II | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-08-30 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1T46
 
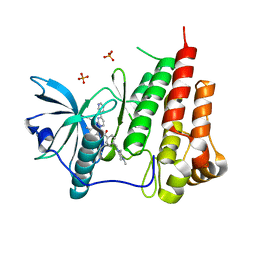 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1T45
 
 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1A3S
 
 | | HUMAN UBC9 | | Descriptor: | UBC9 | | Authors: | Naismith, J.H, Giraud, M. | | Deposit date: | 1998-01-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1ACF
 
 | | ACANTHAMOEBA CASTELLANII PROFILIN IB | | Descriptor: | PROFILIN I | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-07-29 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
