2BKD
 
 | | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | | Descriptor: | Fragile X messenger ribonucleoprotein 1 | | Authors: | Ramos, A, Hollingworth, D, Adinolfi, S, Castets, M, Kelly, G, Frenkiel, T.A, Bardoni, B, Pastore, A. | | Deposit date: | 2005-02-15 | | Release date: | 2006-01-18 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction.
Structure, 14, 2006
|
|
1EKZ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE THIRD DSRBD FROM DROSOPHILA STAUFEN AND A RNA HAIRPIN | | Descriptor: | MATERNAL EFFECT PROTEIN (STAUFEN), STAUFEN DOUBLE-STRANDED RNA BINDING DOMAIN | | Authors: | Ramos, A, Grunert, S, Bycroft, M, St Johnston, D, Varani, G. | | Deposit date: | 2000-03-11 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA recognition by a Staufen double-stranded RNA-binding domain.
EMBO J., 19, 2000
|
|
1IKD
 
 | | ACCEPTOR STEM, NMR, 30 STRUCTURES | | Descriptor: | TRNA ALA ACCEPTOR STEM | | Authors: | Ramos, A, Varani, G. | | Deposit date: | 1996-11-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition.
Nucleic Acids Res., 25, 1997
|
|
6ES4
 
 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
5G1P
 
 | | Aspartate transcarbamoylase domain of human CAD bound to carbamoyl phosphate | | Descriptor: | CAD PROTEIN, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
5G1N
 
 | | Aspartate transcarbamoylase domain of human CAD bound to PALA | | Descriptor: | 1,2-ETHANEDIOL, CAD PROTEIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
5G1O
 
 | | Aspartate transcarbamoylase domain of human CAD in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAD protein, GLYCEROL | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
3COI
 
 | | Crystal structure of p38delta kinase | | Descriptor: | Mitogen-activated protein kinase 13 | | Authors: | Atwell, S, Burley, S.K, Houle, A, Ramos, A, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of p38delta kinase.
To be Published
|
|
2JVZ
 
 | | Solution NMR Structure of the Second and Third KH Domains of KSRP | | Descriptor: | Far upstream element-binding protein 2 | | Authors: | Diaz-Moreno, I, Hollingworth, D, Garcia-Mayoral, M.F, Kelly, G, Cukier, C.D, Ramos, A. | | Deposit date: | 2007-09-28 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Second and Third KH Domains of KSRP
To be Published, 2007
|
|
2BF0
 
 | | crystal structure of the rpr of pcf11 | | Descriptor: | CALCIUM ION, PCF11 | | Authors: | Noble, C.G, Hollingworth, D, Martin, S.R, Adeniran, V.E, Smerdon, S.J, Kelly, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2004-12-02 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key Features of the Interaction between Pcf11 Cid and RNA Polymerase II Ctd.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | Authors: | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-07 | | Method: | SOLUTION NMR | | Cite: | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
5CQU
 
 | | Monoclinic Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
5CQW
 
 | | Tetragonal Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
4B8T
 
 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
2OPV
 
 | |
8USR
 
 | | IL17A homodimer complexed to Compound 23 | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1-[[(1~{S})-1-(8~{a}~{H}-imidazo[1,2-a]pyrimidin-2-yl)ethyl]amino]-1-oxidanylidene-4-phenyl-butan-2-yl]-4,5-bis(chloranyl)-1~{H}-pyrrole-2-carboxamide | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
2N8L
 
 | |
2N8M
 
 | |
2X1A
 
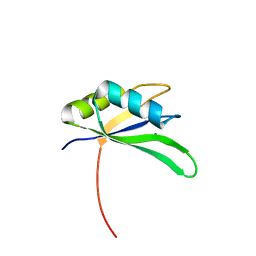 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1B
 
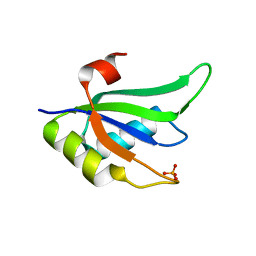 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2JQP
 
 | | NMR structure determination of Bungatoxin from Bungarus candidus (Malayan Krait) | | Descriptor: | Weak toxin 1 | | Authors: | Vivekanandan, S, Jois, S.D, Kini, R.M, Troncone, L.R.P, de Magalhaes, L, Ujikawa, G.Y, Ramos, A.T. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution Structure of Bungatoxin from Bungarus Candidus (Malayan Krait)venom
To be Published
|
|
2KXF
 
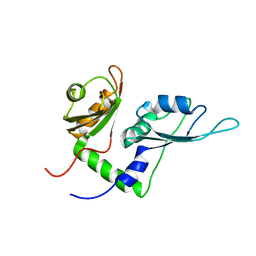 | | Solution structure of the first two RRM domains of FBP-interacting repressor (FIR) | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X1F
 
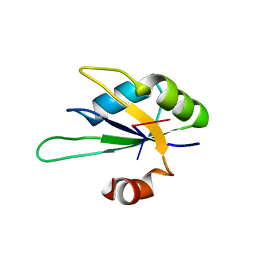 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2KXH
 
 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
